On Cellular Organization and Respiration
-
Singular Plural Kvothe Kvothes Mitochondrion Mitochondria Crista Cristae
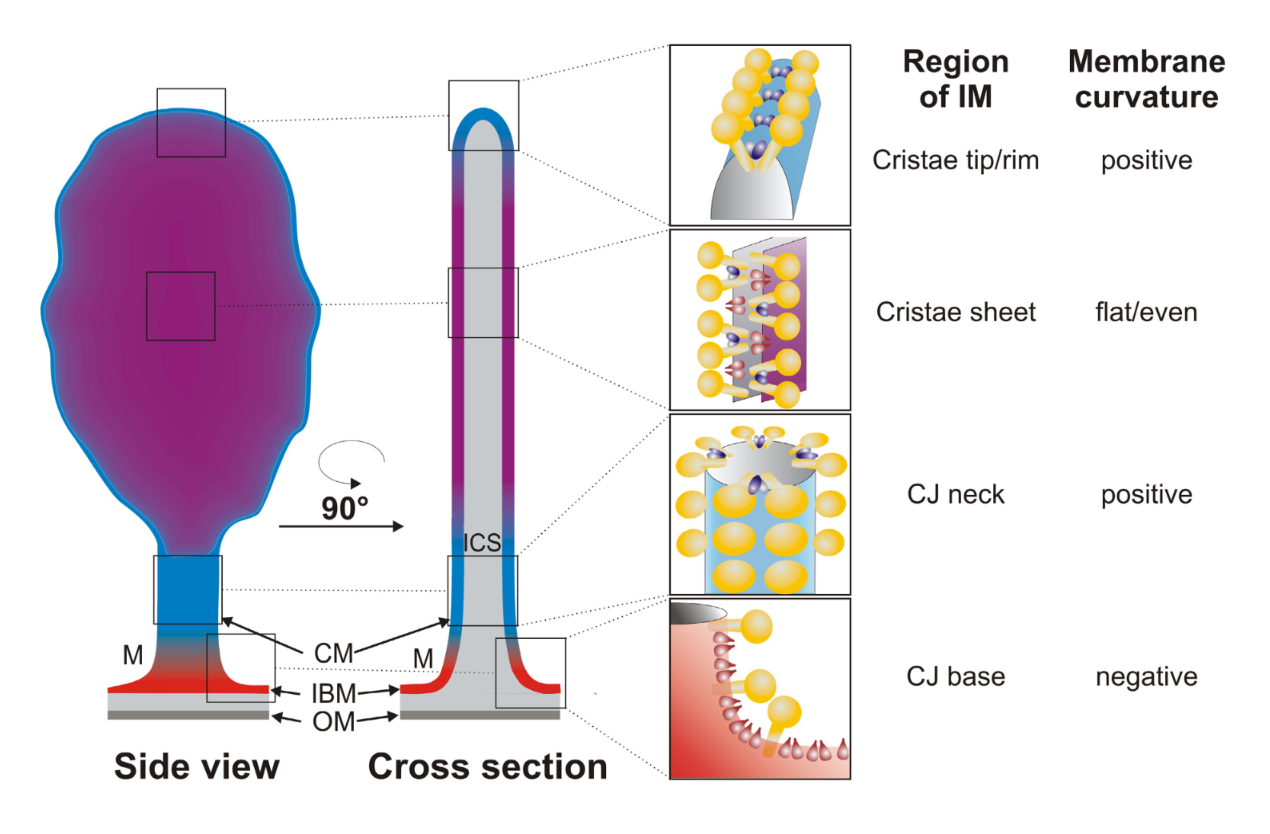
The typical cross section of mitochondria make it seem that their inner membrane is folded as an accordion, but we rarely encounter a rotated view to show that the connection sites can actually be narrow and tubular, and it looks like a deflated balloon.
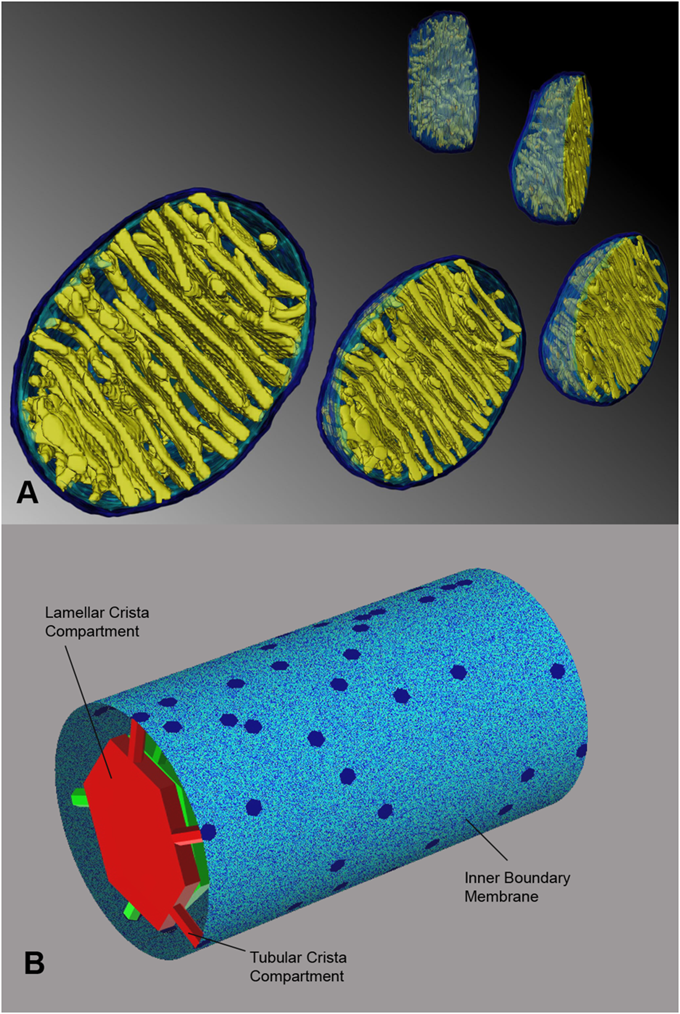
The cristae can expand and shrink depending on respiratory activity.
Anomalous Diffusion Induced by Cristae Geometry in the Inner Mitochondrial Membrane
"It is known since the 1950s, that mitochondria are cylindrically shaped organelles, their silhouette being formed by closely apposing outer and inner membranes, with numerous invaginations, termed cristae within the latter [9], [10]. The cristae forming mitochondrial inner membrane (IM) is contiguous with the peripheral inner membrane, referred to as inner boundary membrane (IBM). Cristae have distinctly different shapes: some organisms and cell types are known to have exclusively tubular cristae formed as curved cylinders of uniform diameter [11], others may exhibit flat or even prismatic cristae. However, in the majority of cells in multicellular organisms, cristae appear as flat membranous infoldings protruding into the mitochondrial body. We will refer to such cristae as lamellar ones. In the 1990s, electron microscopic tomography allowed for more accurate determination of their shape and connectivity to the IBM [12]–[14]. When reconstructed in 3 dimensions (Fig. 1A and figures in [13]), several structural features common to different tissues and species appeared (for a good review see [15]). It was found, that rather than forming folds lamellar cristae should be represented as flattened cisterns of uniform width attached to the IBM with several narrow tubular connections up to hundreds of nanometers long. Attachment places, the junctions, have a diameter (≈28 nm) similar to crista width (≈27 nm). Often, especially in smaller cristae, the flattened part is missing, so that the whole crista consists of the tubular compartment alone. When the flattened segment is present, one crista usually is anchored via several tubular connections. In tissues with large cristae masses, these are densely packed into stacks of parallel cristae, large lamellae mostly oriented perpendicular to the longer mitochondrial axis. In the extreme case of brown adipose tissue tubular compartments are totally missing, but the lamellar parts are still connected to the IBM via short junctions 28 nm in diameter."
A row of ATP synthase pairs in crista formation:
Helical arrays of U-shaped ATP synthase dimers form tubular cristae in ciliate mitochondria
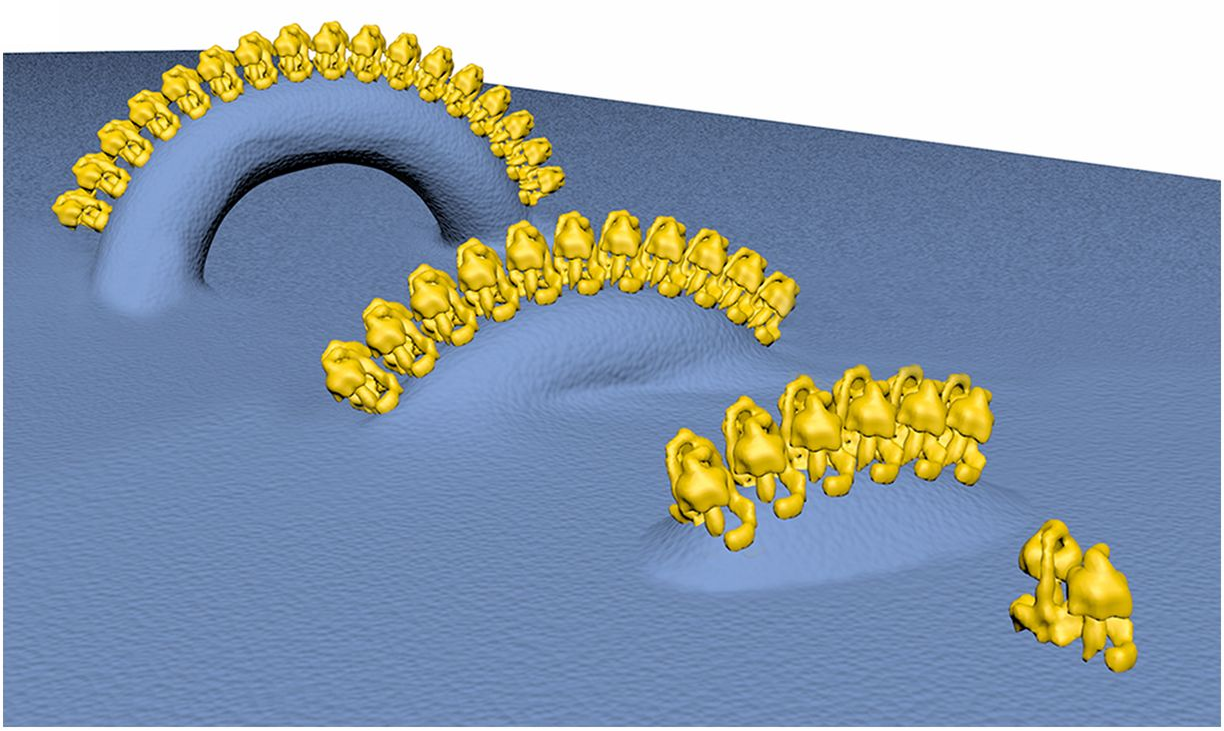
Macromolecular organization of ATP synthase and complex I in whole mitochondria
"The angle between monomers in the dimer was approximately 80° for bovine heart and the three species of fungi, and approximately 115° for potato correlating with the difference observed in F1 head distance of the dimers."
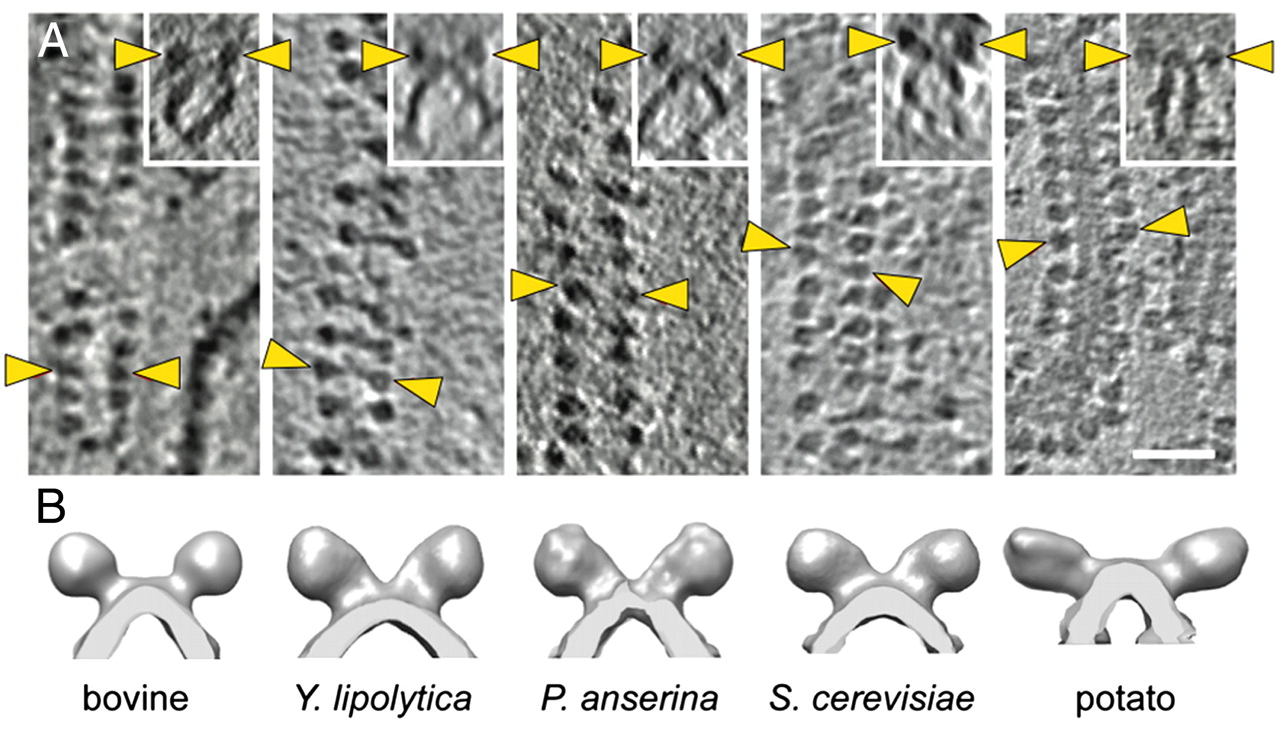
Mitochondrial Cristae: Where Beauty Meets Functionality
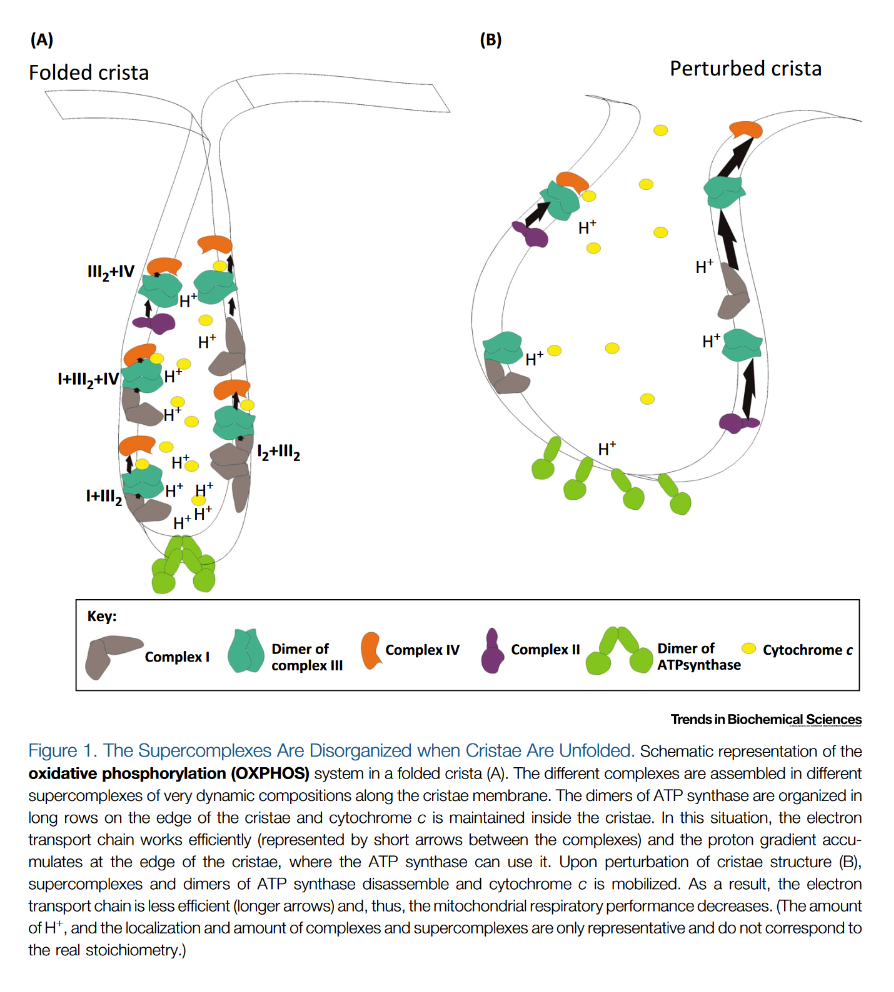
"Cristae are the site of OXPHOS: 94% of complex III and ATP synthase [17] and approximately 85% of total cytochrome c are stored in this compartment [9] (
Box 2)."Respiratory complexes clustering to maximize efficiency.
Macromolecular organization of ATP synthase and complex I in whole mitochondria
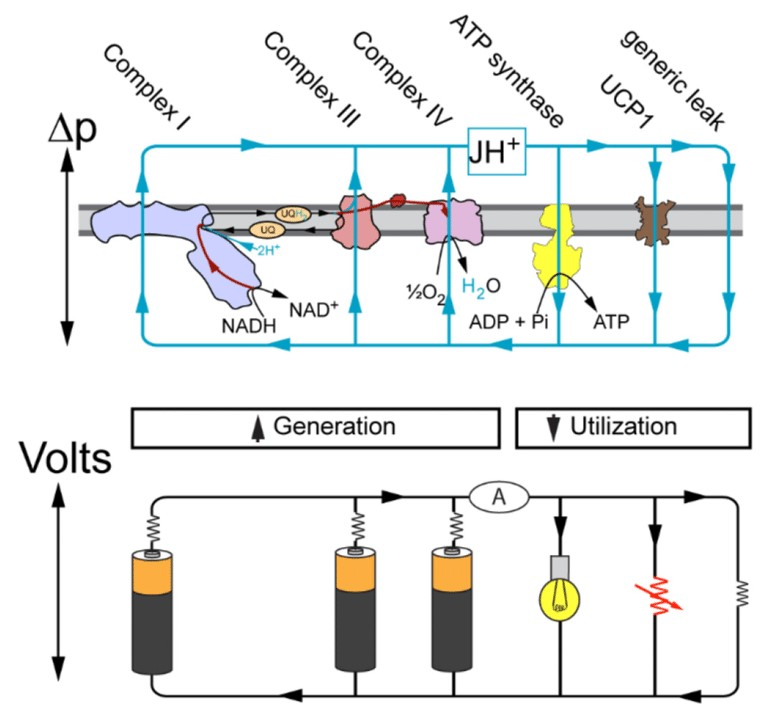
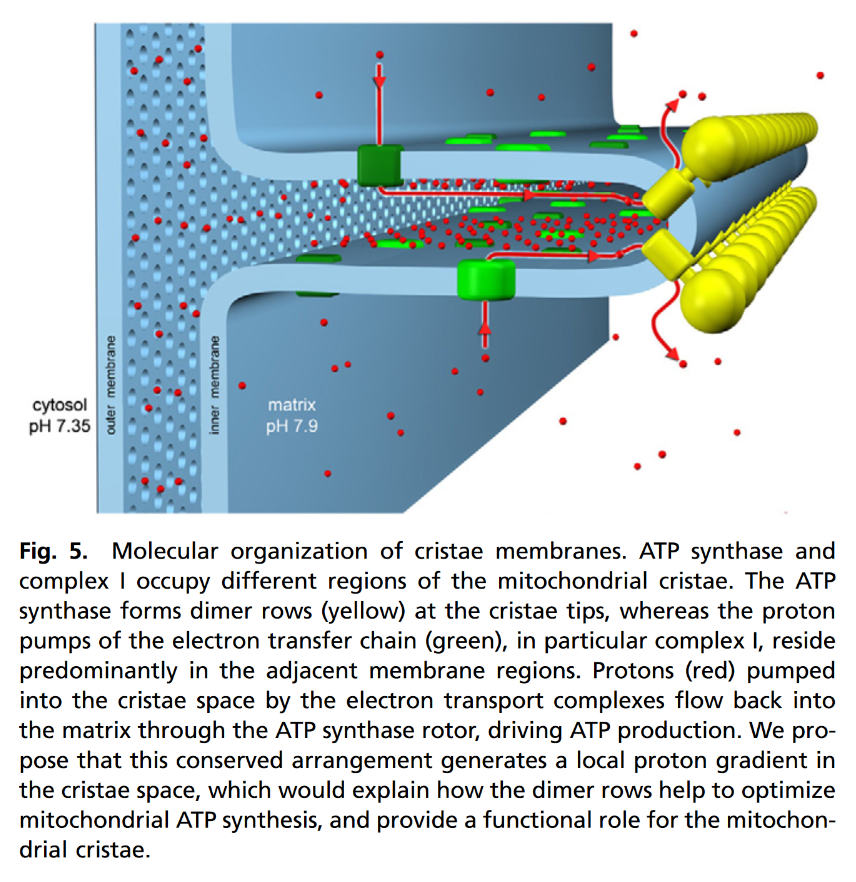
"ATP production in mitochondria is powered by the pmf [proton-motive force], manifest in the electrochemical transmembrane gradient. The resulting current of protons through the Fo rotor of the ATP synthase drives ATP production by rotary catalysis in the F1 head on the matrix side of the membrane. The pmf is generated by complexes I, III, and IV, the proton pumps of the respiratory chain, which translocate protons across the membrane from the matrix side into the cristae space. The pmf has two components, one resulting from the pH difference across the membrane (ΔpH), and the other from the membrane potential (Δψ). In mitochondria and chloroplasts, the pmf is roughly 180 mV. In chloroplasts, this is mostly accounted for by the ΔpH, whereas in mitochondria the ΔpH contribution is comparatively small. Given that the matrix pH is 7.9 (33), the cytosolic pH is 7.35 (34), and the outer membrane is freely permeable to ions, the nominal ΔpH across the mitochondrial inner membrane is 0.55 units, equivalent to 32 mV. However, in vitro studies indicate that the mitochondrial, chloroplast and bacterial ATP synthases all need a ΔpH close to 2 units, equivalent to approximately 120 mV, and a p-side pH close to 6, to produce enough ATP to sustain life (35–38). We propose that the apparent paradox of a ΔpH that is seemingly insufficient and a p-side pH that is too high for efficient ATP production is resolved by the special geometry of the mitochondrial cristae."
"In a previous study (12), we calculated that the high degree of membrane curvature at the cristae ridges [tips] could accommodate a higher charge density for a given membrane potential, resulting in a local ΔpH increase of up to 0.5 units. Furthermore, an earlier modelling study by Cherepanov et al (39) found that protons released on the membrane surface encounter a potential barrier of 0.1 to 0.15 mV. The potential barrier makes it more difficult for the pumped protons to escape to the bulk solvent, resulting in a higher proton concentration, and consequently a lower pH, at distances up to 1 nm from the membrane surface. The effective pH in this boundary layer has been estimated at approximately 6, independent of the pH of the bulk solvent (39). The boundary layer, in combination with the higher charge density at the cristae ridges [at the tips], would meet both the above requirements for efficient ATP synthesis, of a pH below 6 on the p-side of the membrane, and a ΔpH around 2. Protons, which move along membrane surfaces about 10 times faster than into bulk solvent (40), would thus diffuse preferentially along the membrane from their source at the respiratory chain proton pumps on either side of the cristae ridges toward the proton sinks at the ATP synthase dimer rows (Fig. 5). The rapid consumption of protons by the ATP synthases at the cristae ridges would establish a proton gradient along the membrane, by which protons would tend to flow from source to sink, instead of diffusing equally in all directions. The directional proton flow along the membrane surface would provide both a functional role for the mitochondrial cristae, and explain how the conserved, ubiquitous rows of ATP synthase dimers along the cristae ridges help to optimize ATP synthesis in mitochondria."
-
"The first step in glycolysis is the phosphorylation of glucose by hexokinase."
"Phosphorylation of a hexose such as glucose often limits it to a number of intracellular metabolic processes, such as glycolysis or glycogen synthesis. This is because phosphorylated hexoses are charged, and thus more difficult to transport out of a cell."
"Hexokinases I and II can associate physically to the outer surface of the external membrane of mitochondria through specific binding to a porin, or voltage dependent anion channel. This association confers hexokinase direct access to ATP generated by mitochondria, which is one of the two substrates of hexokinase. Mitochondrial hexokinase is highly elevated in rapidly growing malignant tumor cells, with levels up to 200 times higher than normal tissues. Mitochondrially bound hexokinase has been demonstrated to be the driving force for the extremely high glycolytic rates that take place aerobically in tumor cells (the so-called Warburg effect described by Otto Heinrich Warburg in 1930)."
-
@Amazoniac wow, this is fantastic. Thank you so much
-
Flavin adenine dinucleotide | Wikipedia
"Based on the oxidation state, flavins take specific colors when in aqueous solution.
- Flavin-N(5)-oxide (superoxidized) is yellow-orange
- FAD (fully oxidized) is yellow
- FADH (half reduced) is either blue or red based on the pH
- FADH2 (fully reduced form) is colorless"
"90 flavoproteins are encoded in the human genome; about 84% require FAD, and around 16% require FMN, whereas 5 proteins require both to be present. Flavoproteins are mainly located in the mitochondria because of their redox power. Of all flavoproteins, 90% perform redox reactions and the other 10% are transferases, lyases, isomerases, ligases."
"Cellular concentrations of free or non-covalently bound flavins in a variety of cultured mammalian cell lines were reported for FAD (2.2-17.0 amol/cell) and FMN (0.46-3.4 amol/cell)."
Wikipedia has a 'View history' option on the top right corner. You can consult the specific date when something was posted for references.
-
-
Imaginary units ahead:
The Final Frontier of pH and the Undiscovered Country Beyond
"Cells are small. A typical mammalian cell, such as a Chinese Hamster Ovary cell, CHO, has an internal volume of ca. 1.2 pL, of which about 70% is H2O [1]. This corresponds to ca. 2.8×10^13 water molecules inside the cell, calculated by applying Avogadro's number of 6.02×10^23 molecules per mole to the molar concentration of water in water, i.e. 55.56 M. Subcellular compartments are several orders of magnitude smaller than cells. Mitochondria have volumes of ca. 1 fL, (90% in mitochondrial matrix, and 10% in the intermembrane space), lysosomes of about 30 aL, and the smallest of them, coated vesicles, of about 30 to 800 zL (10−21 L) [2]–[4]. In addition, some organelles, such as mitochondria, have variable sizes. They can fuse, divide, and therefore their sizes vary in the range of about an order of magnitude. Thus, in a coated vesicle we find from 7×10^5 to 2×10^7 water molecules, in a lysosome 7×10^8, and in an average mitochondrion 2.3×10^10.
In these organelles, pH values of 6.8 for the mitochondrial intermembrane space, 8.0–8.1 for the matrix, 4–5 for lysosomes, and 5–7 for coated vesicles have been reported [5]. These values have been accepted in the biological community and are hardly ever disputed. In this article, we examine the physical meaning of these pH values."
"Let us consider a mitochondrion, an organelle with a femtomolar volume generating chemical energy for the metabolism of eukaryotic cells [5,8]. This energy is generated by the H+ gradient across the phospholipid bilayer surrounding the mitochondrial matrix (into the matrix). The synthesis of one ATP molecule is accompanied by the transfer of up to three H+ ions along the gradient, from the intermembrane space to the matrix. Thousands of ATP synthase complexes are considered to be simultaneously active [8]. Yet, as shown in
Example 5, simple and obvious calculations demonstrate that there may be no more than seven H+ ions in total in the whole intermembrane space!""Calculations such as those provided in
Examples 5-7indicate that water is too weakly dissociated to be a direct donor or acceptor of H+ and OH− ions at very low biological volumes, even if one can still define the pH formally. It appears that H+ ions can be shuttled to reaction centers by other biomolecules, whose concentrations are not constrained byEq. 2. Best candidates for such molecules are those having pK values close to 7, and are thus capable to serve as donors and acceptors of H+ ions. One obvious group of candidates for such proton donors, available in millimolar and higher concentrations in every intracellular locale are phosphates, e.g. inorganic phosphate, nucleotides and membrane phospholipids.""About 50% of phospholipids carry amine groups with a pK of about 9, indicating that under cellular conditions most of them will be protonated. At this point we end up with a reservoir of over three hundred thousand protons in only just one lysosome! Interestingly, the pK value of the phospholipid amine can be lowered by 1–2 log units, depending on the phospholipid composition of the membrane [12]. Thus phospholipid bilayers may be a significant and tunable source of protons for cellular reactions. It may also the very reason (reason d’etre) of why reactions occur on cell membranes."
"The general view that emerges from the above thought experiments and calculations is that the intracellular pH is in fact a composite of many pairwise proton exchange interactions between individual protonable molecules."
Prefixes applied to the litre | Wikipedia
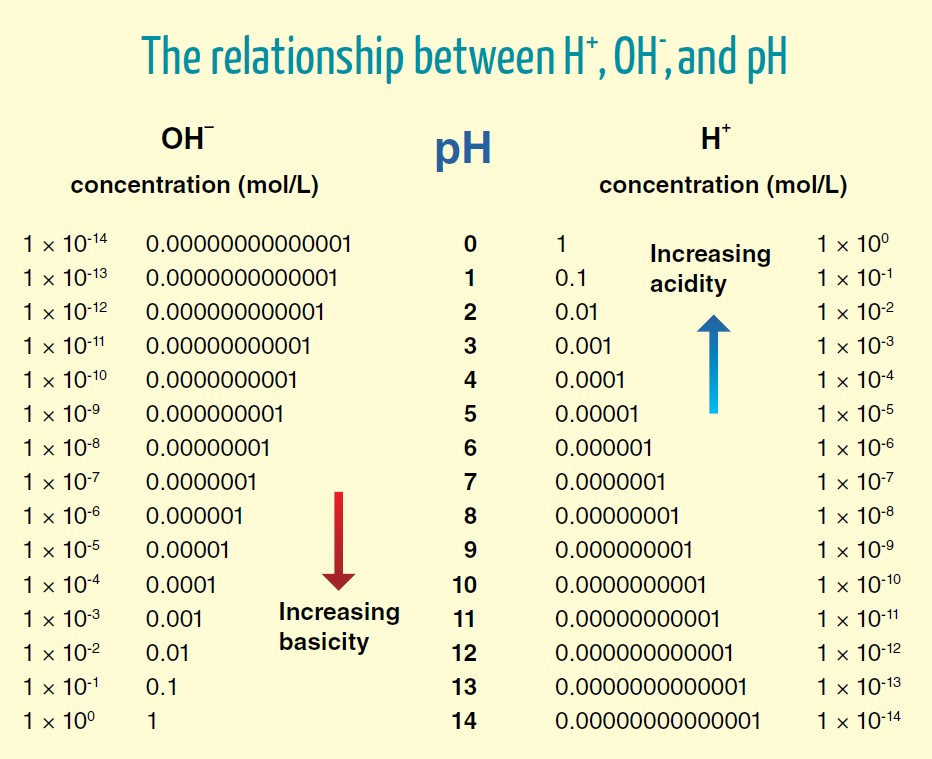
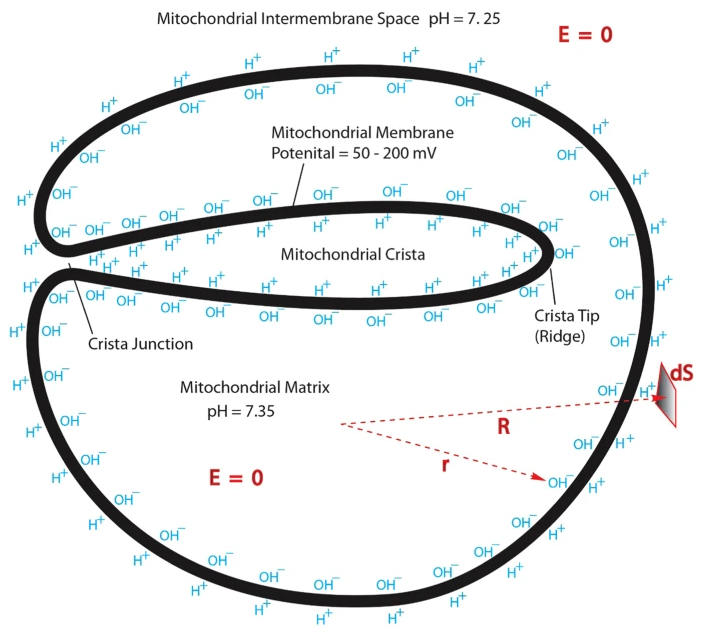
To be generous, we can rely on the pH of 6 proposed above for the region near the surface ('perimembrane') in cristae, that they suggest to be needed for a compartment pH difference of 2 (8–6).
- pH 6 = 0.000001 mol H+/L = 10^–6 mol H+/L
- 1 mol = 6.02 × 10^23 molecules, atoms or ions (H+ in this case)
⠀ - (10^–6) × (6.02 × 10^23) = 6.02 × 10^17 H+
The concentration of protons in cristae with a pH of 6 will be:
- 6.02 × 10^17 H+/L
Mitochondrial volume (as mentioned above):
- 1 fL = 10^–15 L
Water content:
- 70% (or 7 × 10^–1)
Volume distribution:
90% matrix- 10% (or 10^–1) intermembrane space
Therefore:
- 10^–15 L × (70%) × (10%)
- 10^–15 L × (7 × 10^–1) × (10^–1) = 7 × 10^–17 L
The proportional proton quantity:
- 6.02 × 10^17 H+ per 1 L
- ? H+ per 7 × 10^–17 L
⠀ - 6.02 × 10^17 H+/L × (7 × 10^–17 L) ≈ 42 H+ per intermembrane space
Returning to their points that "the synthesis of one ATP molecule is accompanied by the transfer of up to three H+ ions" and "thousands of ATP synthases are simultaneously active", despite the example with a generous value, it's clear that protons must hop between buffers for the current model to be viable.
-
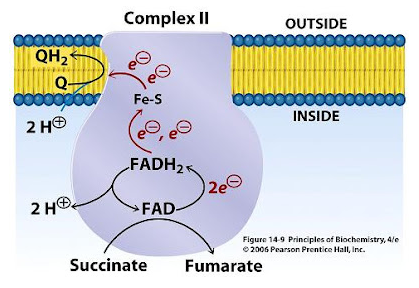
FMN is for Complex I (NADH dehydrogenase) what FAD is for Complex II (succinate dehydrogenase). Both compounds are embedded in their enzyme complex and function as immediate acceptors of electrons from NADH and succinate. This makes NADH oxidation also dependent on riboflavin.
From here.Speaking of fad, despite the association of ketoacid dehydrogenase complexes (such as PDHC↓) with thiamin, they depend on riboflavin as well.
Pyruvate Dehydrogenase Complex
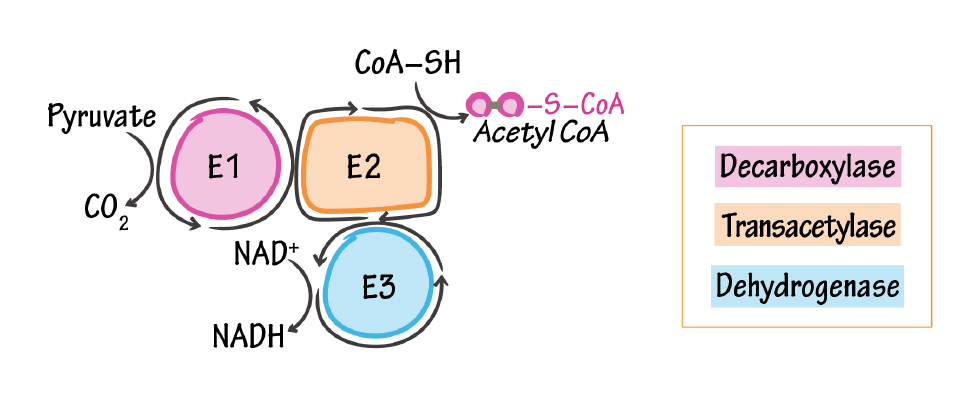
E1, pyruvate dehydrogenase/pyruvate decarboxylase
- Catalyzes pyruvate to acetyl (releases CO2)
- Cofactor: thiamine pyrophosphate (Vitamin B1)
E2, dihydrolipoyl transacetylase
- Attaches CoA to acetyl
- Cofactor: lipoic acid (not vitamin-derived) & coenzyme A (pantothenic acid/vitamin B5)
E3, dihydrolipoyl dehydrogenase
- Reduces NAD+ to NADH
- Cofactor: NAD+ (niacin/vitamin B3) & FAD (riboflavin/vitamin B2)
We have reports of lactate normalization after riboflavin dosing, but it's more challenging to identify where it helped the most, and its function in PDHC may be conserved in deficiency (if I'm not wrong, problems are more common at the decarboxylase part of the complex).
-
ATP Synthase & P:O Ratio | Fundamentals of Biochemistry - UT Austin
Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria
"The most important inference from the presence of the c8-ring in bovine F-ATP synthase, is that 8 protons are translocated across the inner mitochondrial membranes per 360° rotation of its rotor. As each 360° rotation produces 3 ATP molecules from the F1-domain, the bioenergetic cost of the enzyme making an ATP is 2.7 protons."
- 8 H+/3 ATP
- 2.7 H+/1 ATP
"The combination of the electrogenic exchange of internal ATP for an external ADP by the ADP/ATP translocase and the nonelectrogenic symport of phosphate and a proton by the phosphate carrier protein adds one proton to the total required to provide ATP to the cellular cytoplasm, and so the bioenergetic cost to the vertebrate mitochondrion is 3.7 protons."
- 3.7 H+/1 ATP (extra H+)
"Where they are known, the sequences of c-subunits are identical in almost all vertebrates (
Fig. 4). Also, they are highly conserved across animal phyla (Fig. 5), and mutations are confined mostly to the N and C termini of the protein, away from the c-ring-central stalk interface, and the ion binding site around Glu-47." "Therefore, it appears that F-ATPases in all vertebrates and probably all or most invertebrates, will contain c8-rings. Therefore, the bioenergetic cost of the enzyme making an ATP is 2.7 protons in vertebrates and probably in invertebrates also (exceptions dictated by particular energetic demands of a species may exist). There are estimated to be 48–58,000 vertebrate species and about two million invertebrates. This picture of an evidently universal cost of making an ATP molecule in multicellular animals is very different from the picture in prokarya, chloroplasts, and fungi where, for reasons that are not understood, the F-ATP synthases have evolved a range of ring sizes from c10–c15 with associated higher bioenergetic costs of 3.3–5 protons per ATP (22). Thus, the vertebrate and probably the invertebrate F-ATP synthases are the most efficient that have been found.""In mitochondria, it is thought that for each two electrons transferred to oxygen from NADH or succinate, 10 or 6 protons, respectively, are pumped out of the matrix into the space between the outer and inner membranes of the organelle."
- 10 H+/NADH [4 (C-I) + 4 (C-III) + 2 (C-IV)]
- 6 H+/succinate [
4 (C-I) +0 (C-II) + 4 (C-III) + 2 (C-IV)]
"Therefore, the number of moles of ADP phosphorylated to ATP per two electrons transferred to oxygen, known as the P/O ratio, will be 10/3.7 and 6/3.7, or 2.7 and 1.6 for NADH and succinate, respectively, close to experimental values of 2.5 and 1.5 (23)."
- 3.7 H+/1 ATP (⇈ with the extra proton)
- 1 H+/0.27 ATP
NADH and succinate are 2-electron donors despite their difference in moving protons. The oxygen (½O2) consumption brought up in the previous paragraph represents the oxidation of NADH or succinate.
- 1 H+ (from a generic source) → 0.27 ATP
- 10 H+ (from 1 NADH) → 2.7 ATP/NADH ≈ 2.5 ATP/NADH
- 6 H+ (from 1 succinate) → 1.6 ATP/succinate ≈ 1.5 ATP/succinate or FADH2
Complete glucose oxidation yields more than 1 NADH and 1 FADH2 molecule. It can be simplified as:
Cytosol
Glycolysis
- 2 NADH (GAPDH)
- 2 ATP
Mitochondria
Pyruvate oxidation
- 2 NADH (PDH)
- 2 CO2
--
Tricrapoxylate cycle
- 3 NADH (IDH, KGDH, MDH)
- 1 FADH2 (SDH)
- 1
GTPATP - 2 CO2
Tricrapoxylate cycle ×2
- 6 NADH
- 2 FADH2
- 2
GTPATP - 4 CO2
Cytosol + Mitochondria:
- 10 NADH
- 2 FADH2
- 4 ATP ('substrate-level phosphorylation')
- 6 CO2
Assuming that (former section):
- 2.5 ATP/1 NADH
- 1.5 ATP/1 FADH2
We have:
- 10 NADH → 25 ATP (oxidative phoshorylation)
- 2 FADH2 → 3 ATP (oxidative phoshorylation)
- 4 ATP ('substrate-level' phosphorylation)
- 6 CO2
Combined:
- 28 ATP (oxidative phoshorylation)
- 4 ATP ('substrate-level' phosphorylation)
- 6 CO2
Grand total per complete glucose oxidation:
- 32 ATP
- 6 CO2
Out of 32 ATP, 2 are the net cytosolic yield and 30 are mitochondrial.
Most of the ATP is produced in mitochondria, but consumed in the cytosol. When the phosphorus-containing molecules are returned to the mitochondria to reform ATP, they pass through the intermembrane space and allegedly dissipate the extra proton quantified above.
However, if protons concentrate in cristae, which in turn are relatively isolated compartments, ADP and phosphate imported from the cytosol might not interfere with this pool of protons.
-
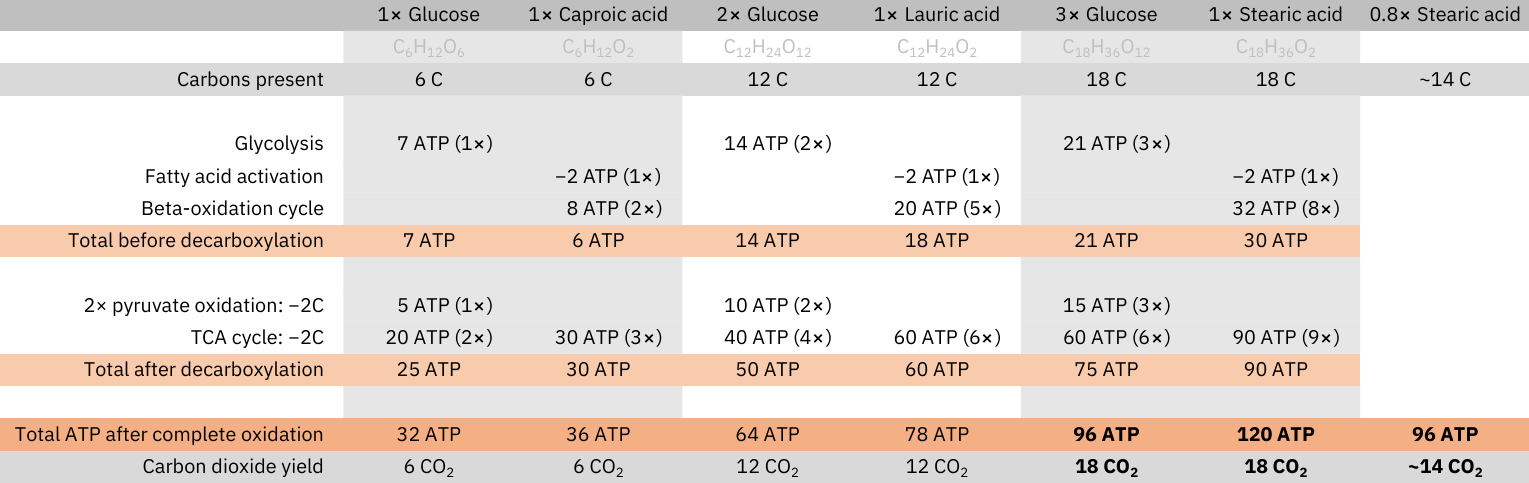
The complete oxidation of glucose doesn't produce more CO2 than that of fatty acids: every carbon that composes these molecules is eliminated as CO2 (decarboxylation), and at no stage in their oxidation carbons are added.
Yield of complete oxidation (matched by carbons):
- 1 glucose (C6H12O6): 6 CO2 and 32 ATP
- 1 caproic acid (C6H12O2): 6 CO2 and 36 ATP
⠀ - 2 glucose (C12H24O12): 12 CO2 and 64 ATP
- 1 lauric acid (C12H24O2): 12 CO2 and 78 ATP
⠀ - 3 glucose (C18H36O18): 18 CO2 and 96 ATP
- 1 stearic acid (C18H36O2): 18 CO2 and 120 ATP
Since fatty acids produce more energy per carbon than glucose, for a given energetic need, meeting it through fatty acid oxidation is economical, needing less carbons to metabolize, and less exposure to CO2.
Fatty acids oxidation is more dependent on FAD relative to glucose oxidation (needed for ACAD enzyme in every normal beta-oxidation cycle). Taken together with the previous information..
- 2.5 ATP/NADH
- 1.5 ATP/FADH2
..it can give a false impression that fatty acids oxidation must produce less energy, but the simple comparison above is enough to conflict with this.
A major factor responsible for the lower ATP yield in glucose oxidation is pyruvate dehydrogenase, for eliminating carbons (
2/61/3 of them) before they enter the TCA cycle.- 2 carbons at PDH → 2 NADH + 2 CO2 → 5 ATP + 2 CO2
- 2 carbons at TCA cycle → 3 NADH + 1 FADH2 + ATP + 2 CO2 → 10 ATP + 2 CO2
The carbons of fatty acids are all eliminated in the TCA cycle.
In addition, (1) the last acetyl-CoA formed in beta-oxidation is a by-product of the final cycle, and it dispenses oxidation along with the respective reduced coenzymes of each cycle. (2) Fatty acids also undergo activation with the consumption of what's equivalent to 2 ATP per molecule.
The two factors just described counteract the beta-oxidation advantage:
- Glycolysis: ~2.3 ATP/2 C
- Beta-oxidation: 4 ATP/2 C
When the fatty acids chains are shorter, the ATP yield per carbon before decarboxylation is similar between fatty acids and glucose (as can be verified in the table), but as the fatty acids chains increase in length, the counteracting factors are minimized for only affecting a lesser and lesser portion of the chain, making the greater ATP yield from beta-oxidation (products) stand out.
-
7.2 - Complex I | Structure & Reactivity in Organic, Biological and Inorganic Chemistry (Schaller)
"The main events in Complex I are summarised in the cartoon below. You can see the electrons entering from the matrix at the bottom of the picture (the pathway is shown by the blue arrows). They are delivered by NADH and handed off to FMN; this step will be discussed below. The electrons are transferred via outer sphere electron transfer through a series of iron sulfur clusters and are eventually delivered to the lipid-soluble ubiquinone (Q)."
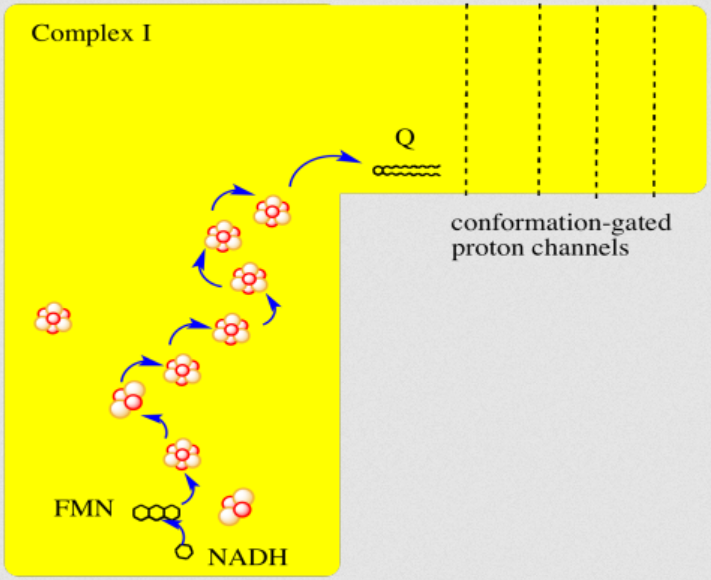
"By far the most common electron acceptor in the oxidative phosphorylation supercomplex is an iron atom. Of course, the most common oxidation states for iron ions are Fe2+ and Fe3+. An iron in the 3+ oxidation state is able to accept an electron, becoming Fe2+. In contrast, an iron in the 2+ oxidation state could pass an electron on, becoming Fe3+ in the process."
"NADH is a two electron donor. An Fe3+ ion is a one-electron acceptor. We need an adapter for this electrical connection. The adapter comes in the form of FMN. FMN is the structure with some atoms coloured in blue and red near the bottom right corner of the picture.
- NADH only donates two electrons at a time.
- The iron ions in the electron transport chain can toggle between Fe(III) and Fe(II); they can accept only one electron at a time.
- An adapter is needed to convert two-electron transfer into one-electron transfer.
FMNH2 is a little bit like NADH. Its oxidized form, FMN, can accept two electrons and a proton in the form of a hydride ion, as well as an additional proton. In other words, FMN accepts H– and H+ to become FMNH2. However, a slightly different route is available to FMN. It can also undergo reduction one electron at a time. In reality, the addition of an electron would be shortly preceded by or shortly followed by addition of a proton, in order to keep the overall charge the same. This state, FMNH, is called the semiquinone form.
What's the difference between NAD and FMN? Why is one able to accept only a pair of electrons, whereas the other can accept one at a time? When FMN accepts one electron, it becomes a radical. Radicals are unstable, reactive species. They can be stabilised chiefly by delocalisation. The additional conjugation in FMN compared to NAD allows the odd electron to be delocalised more extensively in FMN. That radical stability is the key difference.
- The presence of extended conjugation stabilises a radical on FMNH.
- The stability of this radical allows FMN to accept one electron at a time.
Once FMNH2 has formed, the reverse is true, of course. It can give up one electron at a time. As a result, FMN can take a pair of electrons coming in from NADH and send them out one at a time into the electron transport chain."
7.3 - Complex II | Structure & Reactivity in Organic, Biological and Inorganic Chemistry (Schaller)
"In terms of how those electrons make their arrival, the picture is somewhat similar to Complex I. Succinate is a hydride donor in this reaction, so it is donating two electrons. As in Complex I, a series of FeS clusters will eventually relay the electrons, so we have a matching problem. To step down from a two-electron process to a one-electron process, another flavenoid, FAD, acts as an intermediary. Like the related FMN, FAD can accept either one or two electrons at a time, and its reduced form, FADH2, can donate either one or two electrons at a time.
- Just like in Complex I, an adapter is needed to convert the two-electron process of succinate oxidation to the one-electron process of iron reduction.
- The adapter is another flavenoid structure, FAD, which is very similar to the FMN used in Complex I.
After the electrons have arrived and are stored in the form of FADH2, they are passed along, one at a time, through a series of three FeS clusters. Just as in Complex I, these carriers are all bound to the proteins, so they are held in one place and do not move. The outer sphere mechanism by which the electrons are transferred can only operate over a short distance, from one electron carrier to the next."
-
NADPH Oxidases (NOX): An Overview from Discovery, Molecular Mechanisms to Physiology and Pathology
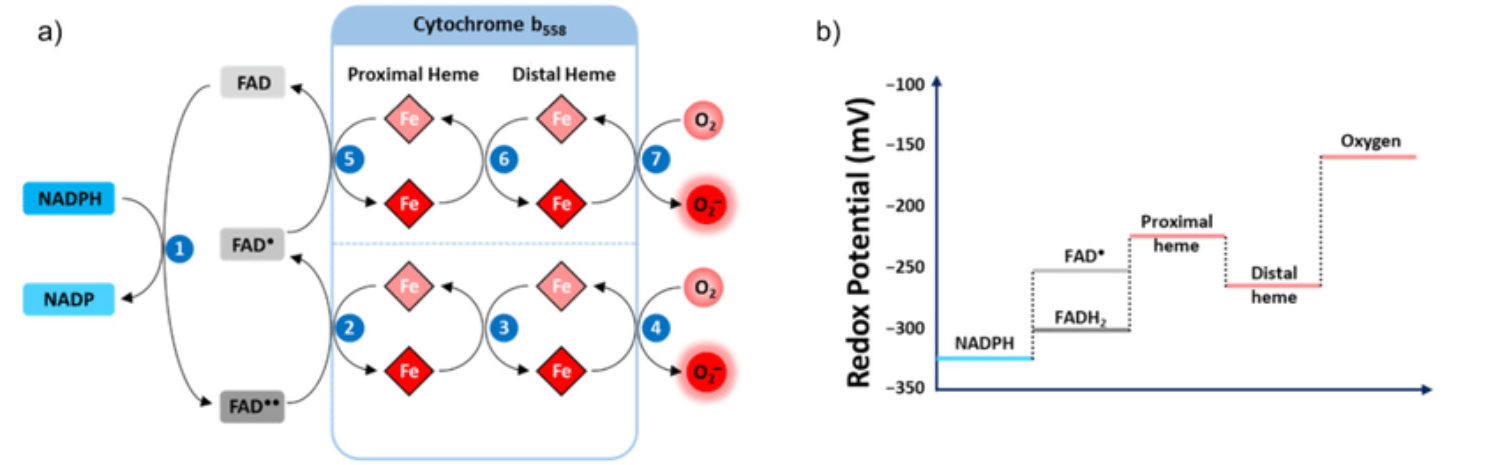
This relates to the previous post and to the next information.
If you want to question your current understanding of cellular respiration, check out the work of Kelath Manoj and his subversive murburn model (stands for 'mild, unrestricted burning'), where he challenges anything that crosses the way.
- Murburn concept | Wikipedia
- Kelath Manoj | ResearchGate
- Kelath Manoj | Google Scholar
- Satyamjayatu (his foundation)
- Satyamjayatu | YouTube
⠀ - Murburn concept: A facile explanation for oxygen-centered cellular respiration
- Mitochondrial oxidative phosphorylation: Debunking the concepts of electron transport chain, proton pumps, chemiosmosis and rotary ATP synthesis
- Murburn concept and murzymes in 2023: Celebrating 25th year of pursuit
He treats (diffusible) reactive oxygen species (DROS) as an essential part of cellular respiration. Speculates that the projections of respiratory complexes were evolutionarily shaped to take advantage of radical generation. He identifies sites of ATP synthesis in all respiratory complexes and it's as oxidative phosphorylative as it gets: a reduction of oxygen to superoxide in respiratory complexes leads to its diffusion towards local ADP or phosphate attack, and a straightforward reaction joins them to form ATP without involving the convoluted steps of protons movement to drive ATP synthase (Complex V) subunit rotation.
- Superoxide radical as electron donor for oxidative phosphorylation of ADP (referenced by him)
Aerobic respiration: Proof of concept for the oxygen-centric murburn perspective
A respirawesome or supercomplex [I(III)2IV] showing 11 ADP-binding sites within projections into the matrix:
- 6 in Complex I
- 4 in Complex III
- 1 in Complex IV
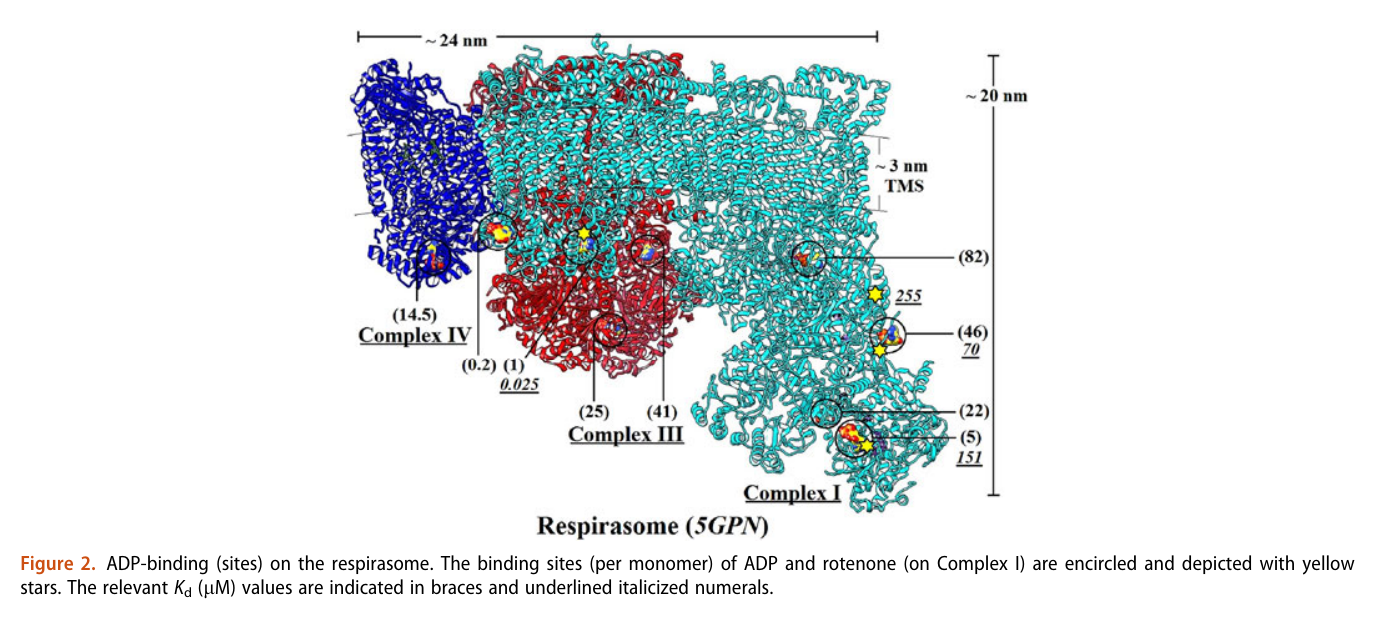
His team also report 'cavities or channels' that promote diffusion of oxygen species to desired targets (be them ADP, phosphorus or the known electron acceptors):
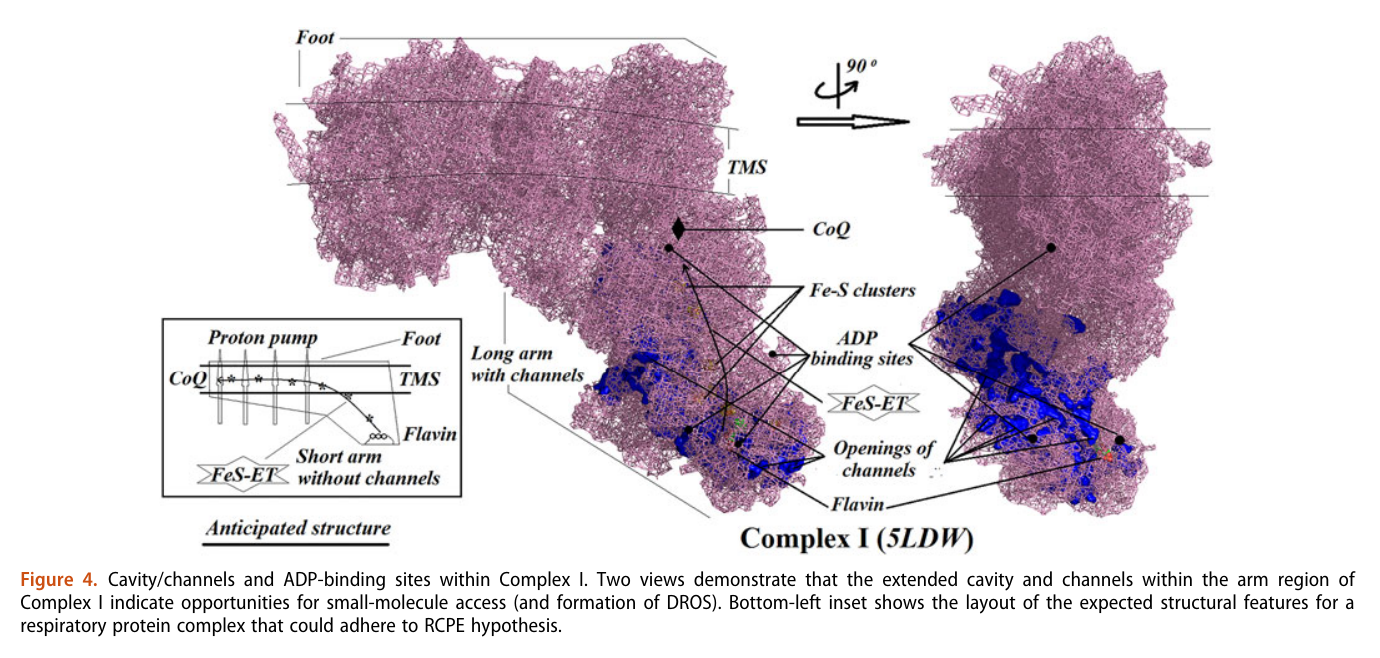
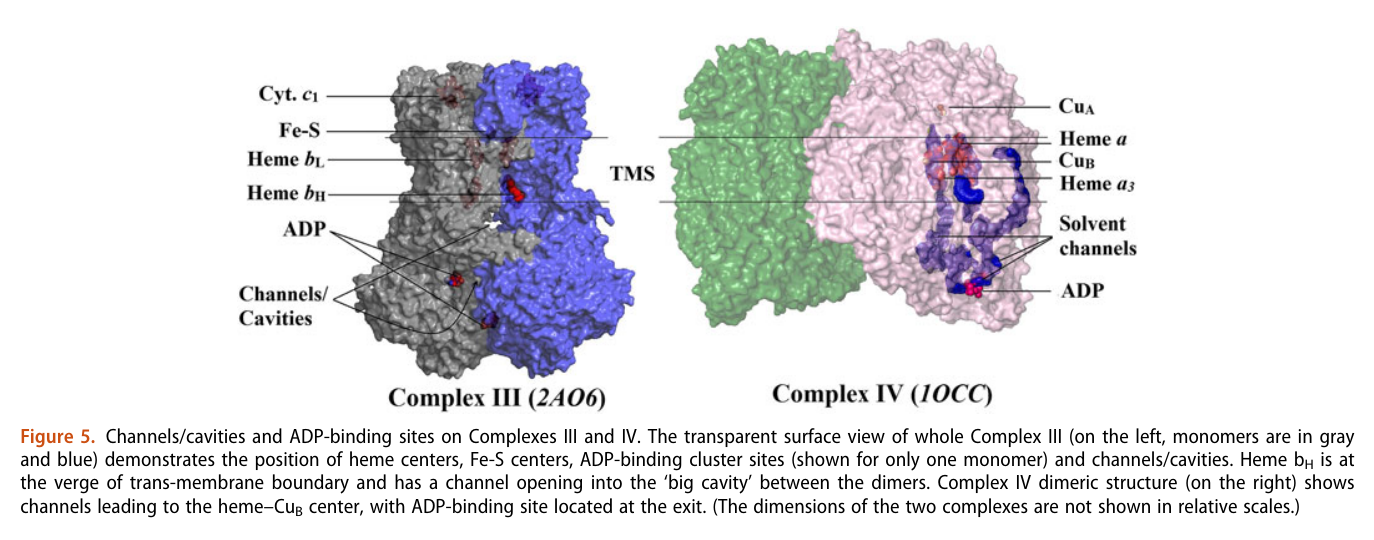
The proposed chain of reactions is shown here (omitting what occurs in between each):
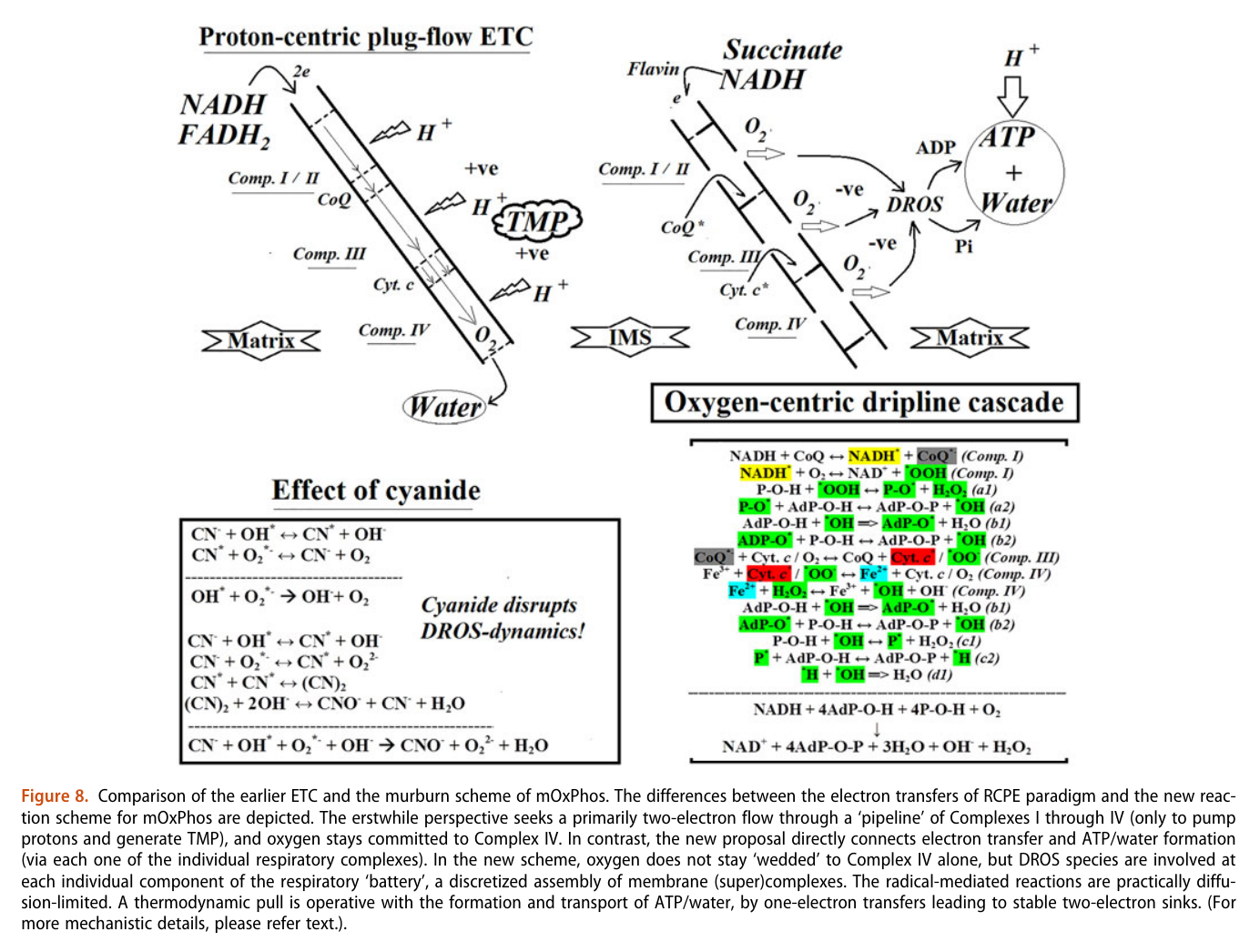
AdP-O-H ADP P-O-H Pi (inorganic phosphate) AdP-O-P ATP When complexes cluster, the organization favors their interaction, and ADP-binding sites from one complex can make use of diffusing reactive oxygen species from another. The strategic location and shared metabolites are shown in this diagram:
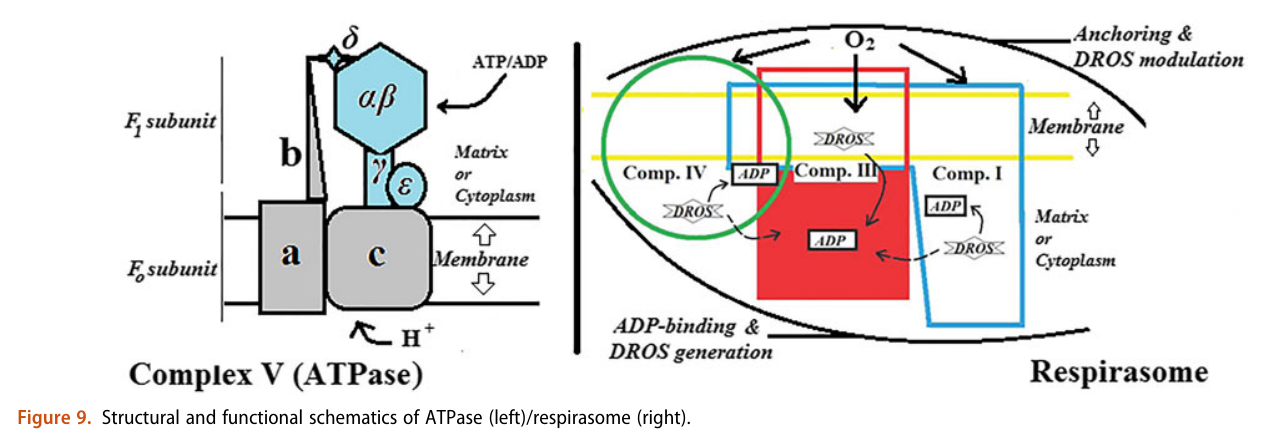
He not only diminishes the importance of Complex V as a source of ATP synthesis, but suggests that it's a decomposer of ATP (an ATPase). According to him, Complex V has much higher affinity for ATP than ADP, which is odd for a protein that's intended to produce ATP. To compound the issue, he argues that ADP is available at lower concentration than ATP.
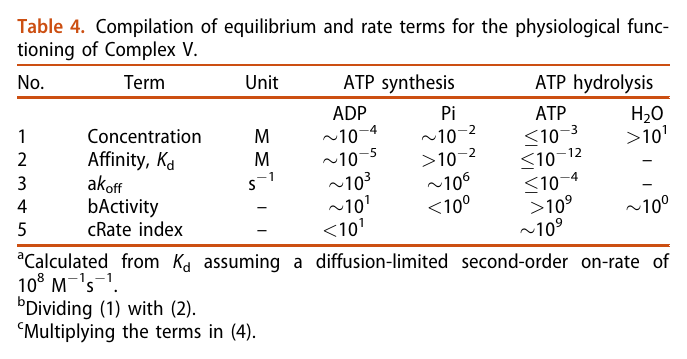
The half-affinity for ADP being close to its concentration would be a problem.
He acknowledges the meek quantity of free protons per mitochondrion, which would be another concern for a 'rotary' protein dependent on them, but rejects the buffering or hopping possibility. I haven't looked into why. He adds to the picture the following considerations:
- Complexes per mitochondrion: ~10^4-10^5
- Proton requirement: ~10^6 (which is ×10 the previous value, assuming 10 H+ extruded per 1 NADH oxidized in the traditional model)
There's a lot more to uncover from his work. As an example, he's familiar with Gilbert Ling's model, compares it with his proposal and the traditional approach.
-
A while ago I came across this publication through GembaRed's articles:
However, Tony questioned its validity:
Human blood contains circulating cell-free mitochondria, but are they really functional?
"As noted by Bertero et al. (7), the evidence provided by Dache et al. (4 ⇈) was not sufficient to assess the functionality or the respiratory competence of human circulating cell-free mitochondria. By conducting a comprehensive analysis of mitochondrial bioenergetics of human circulating cell-free mitochondria in direct comparison with mitochondria extracted from platelets of the same blood samples, we found no significant support for the functionality or the respiratory competence of those cell-free mitochondria. Indeed, there was no significant evidence that cell-free mitochondria possess a functional electron transport system since they exhibited no responses to mitochondrial substrates, ADP, or inhibitors. Yet, when fueling directly complex IV with exogenous electrons (i.e., Asc + TMPD), a significant mitochondrial O2 consumption was detected [similarly to Dache et al. (4), except that no negative control was provided in their study]."
"Although fully rejecting the hypothesis that human circulating cell-free mitochondria are functional would require a way larger sample size (e.g., n > 90 for reaching a statistical power of 0.80 for OXPHOSCI + II based on data presented in Fig. 1B), our results do not support the functionality of circulating cell-free mitochondria. Further studies investigating the functionality or lack of functionality of circulating cell-free mitochondria are now required, since this appears as a critical point to elucidate their potential physiological role(s) [e.g., (8)]."
-
NAD is used here as the generic term for NAD+ and NADH. Despite their reactive site in evidence, both have an overall negative charge because of the phosphate groups.
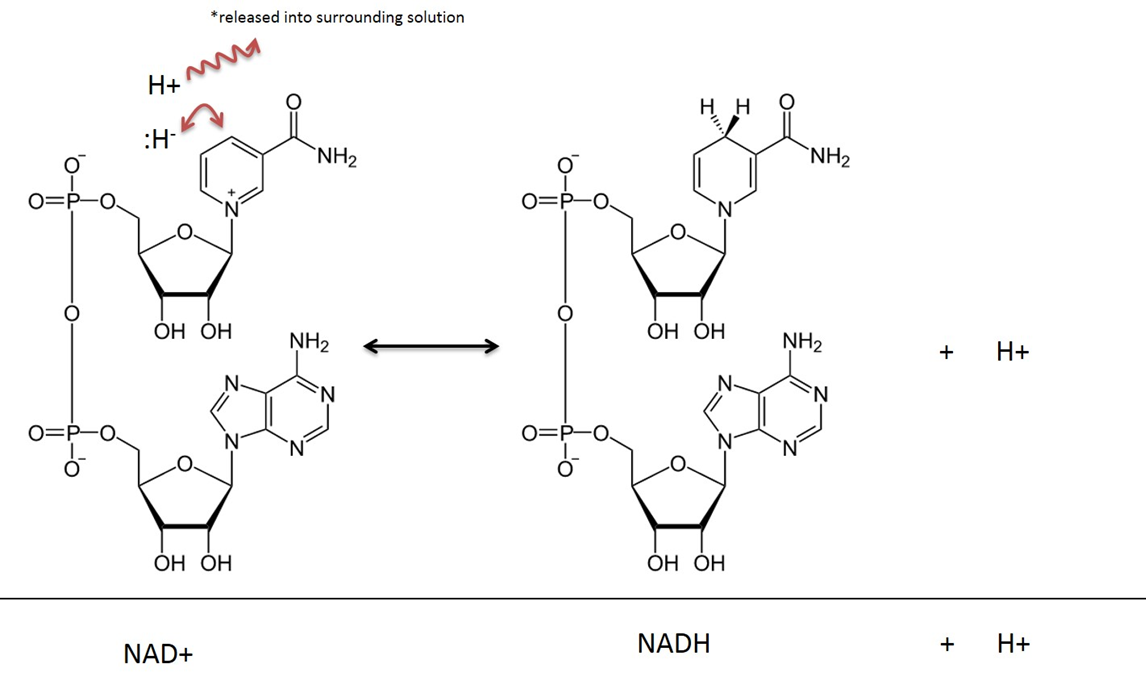
In addition to being charged molecules, they're bulky.
These factors make mitochondrial matrix unable to exchange NAD with the cytosol. For similar reasons, glucose is trapped once inside the cell after phosphorylation by hexokinase.
NADH within the matrix is oxidized locally and the electrons are channeled to oxygen. However, a fraction of NADH is formed in the cytosol (GAPDH). This NADH has to be oxidized as well, to regenerate NAD+ for glycolysis to continue.
The inability to import cytosolic NADH into the matrix is circumvented by the known mitochondrial shuttles. NADH can transfer its electrons to a temporary carrier, one that's importable, and the electrons (with protons) can be recovered once inside.
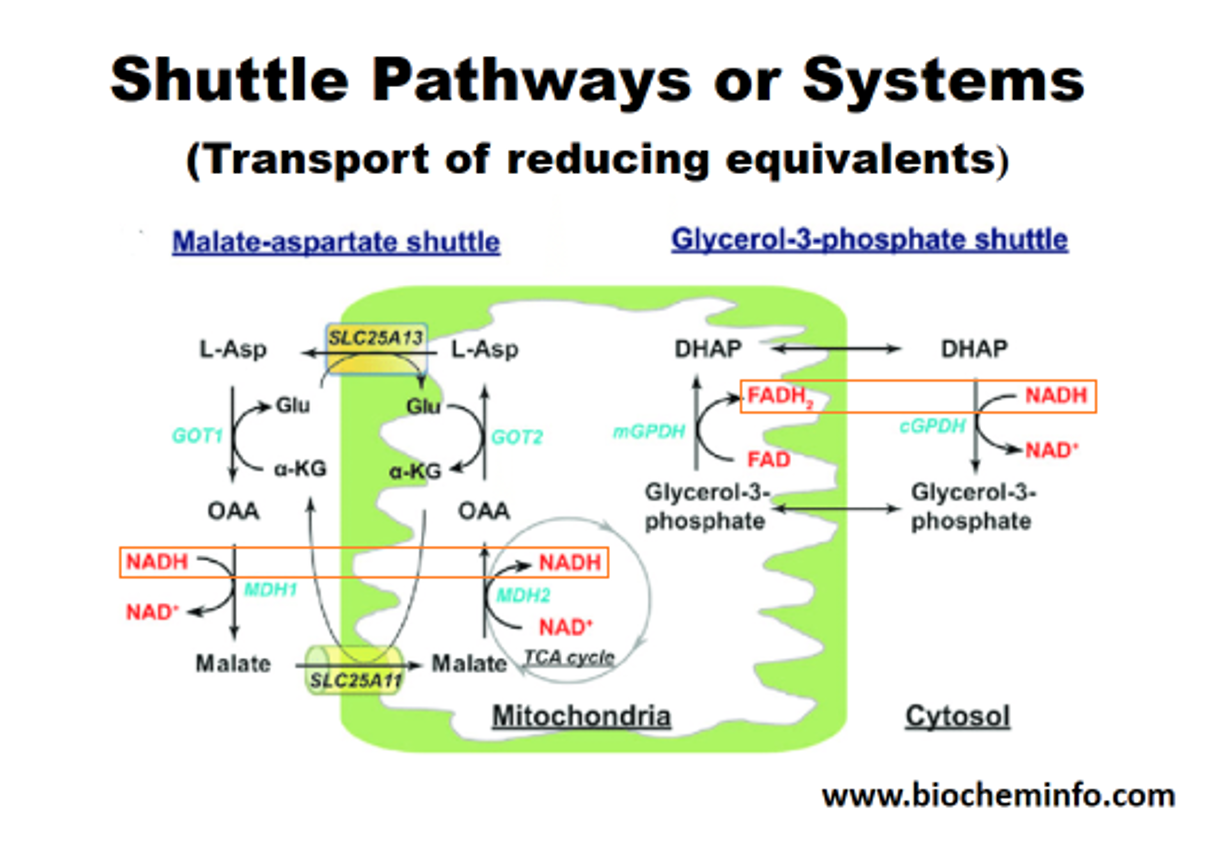
The method on the left appears to be the predominant in tissues with high energy demands, although both can be important. In support:
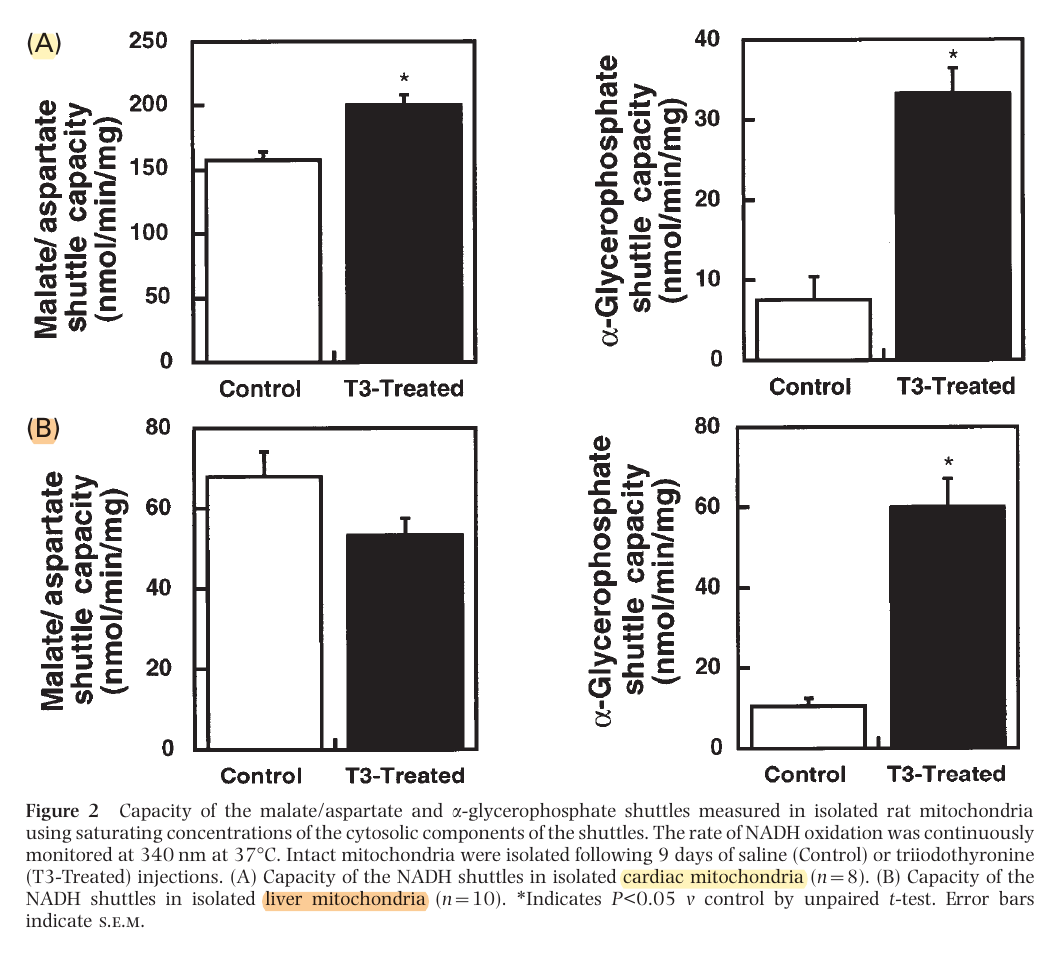
This way, most of the NADH is reformed on the matrix side. Or else, transferring the electrons to FAD would result in less energy production, whether we go by the traditional model (10 H+/NADH versus 6 H+/FADH2) or Kekel's approach.
The external layer of the mitochondria is much more permeable than the internal. For the specific location of mGPDH:
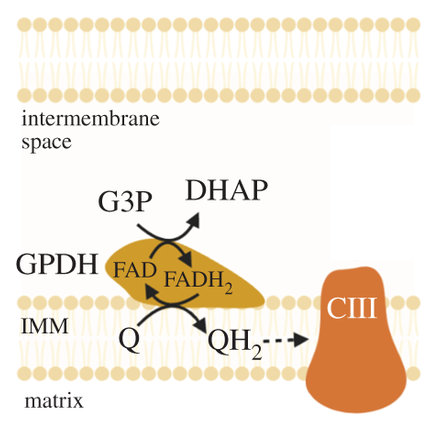
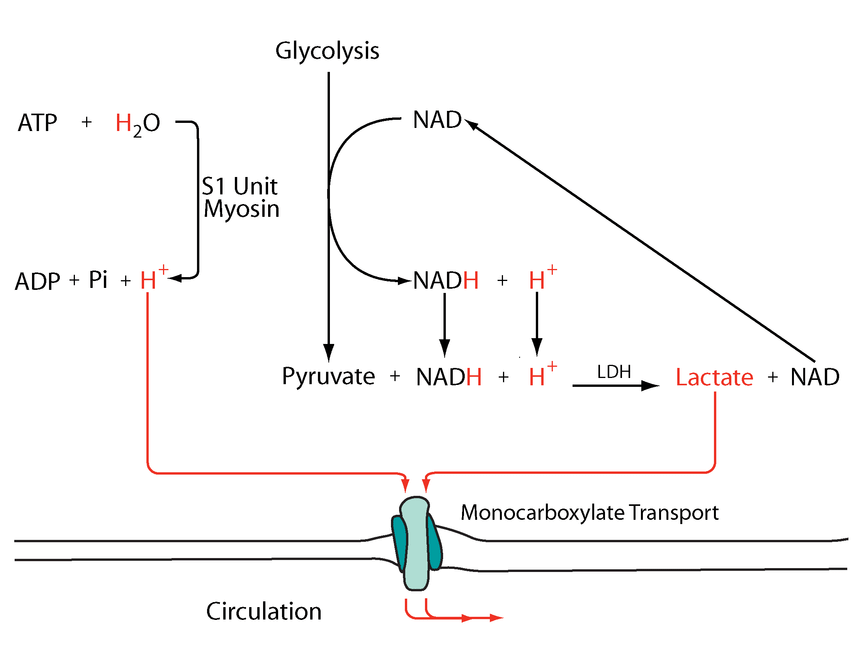
But prior to crossing comparments, pyruvate and NADH are in equilibrium with lactate and NAD+.
Mitochondrial lactate metabolism: history and implications for exercise and disease
Keq (equilibrium constant) = [Lactate][NAD+] / [Pyruvate][NADH]
A high ratio of lactate to pyruvate is a normal occurrence in tissues. We have authors who go to the extent of treating lactate as the final product of healthy glucose breakdown in the cytosol (but acknowledge the problems when elevated in disease).
- Lactate, Not Pyruvate, Is the End Product of Glucose Metabolism via Glycolysis
- Lactate is always the end product of glycolysis
Lactate:pyruvate Tissue Error Kvothean 7:1 Liver 13:1 Skeletal muscle (rest) 23:1 Brian (not to be confused with brain) ≥25:1 Traumatic brian injury 159:1 Skeletal muscle (immediately after exhaustive exercise) A generic ratio would be lactate 10:1 pyruvate.
⠀
NAD+/NADH and skeletal muscle mitochondrial adaptations to exerciseCytosol
- [NAD+] = 0.15 mM
- [NADH] = 0.00028 mM
- NAD+ ~540:1 NADH
Mitochondrial Matrix
- [NAD+] = 3.15 mM
- [NADH] = 0.5 mM
- NAD+ ~6:1 NADH
It's an advantage to not have NAD moving freely between compartments to maintain the desired ratios in each.
From the information above, an example with skeletal muscle (assuming cytosolic ratios for both):
Lactate 13:1 pyruvate NAD+ 540:1 NADH (Keq = 7020)
Therefore, the favored molecules are products of the LDH reaction in one direction.
- Pyruvate + NADH (+ H+) -(LDH)→ Lactate + NAD+
Lactate dehydrogenase has 'very high activity', that 'exceeds glycolytic capacity', which contributes to their proportional variations respecting the ratio in the given tissue, possibly under great influence of NAD. If pyruvate and NADH levels increase, a corresponding increase in lactate and NAD+ tends to follow.
Through lactate formation, NAD+ can be regenerated without depending on the matrix (glycolysis can function independently from oxidative phosphorylation), and this lactate can be distributed to other cells if it can't be metabolized locally. It's a feature that's advantageous for cancer cells.
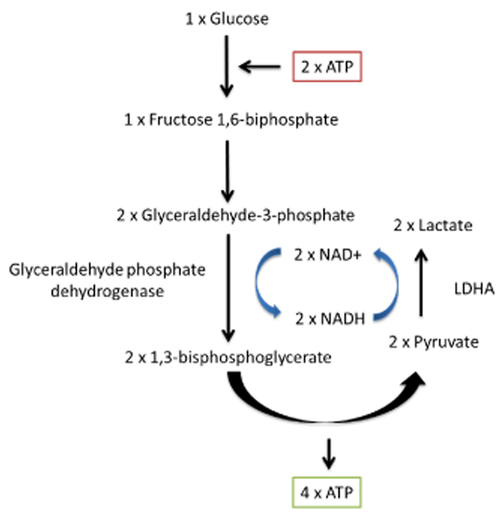
You'll find two lactate shuttles being discussed by researchers:
- Intercellular (cell-to-cell)
- Intracellular (cytosol-to-matrix)
The redistribution of lactate between organs is a well-known phenomenon, hence the lactate (Cori) cycle. It's a wasteful cycle because glycolysis net-produces 2 ATP and 'reverse glycolysis' (gluconeogenesis) consumes 6 ATP. But lactate doesn't have to undergo reverse glycolysis to reform glucose elsewhere, to then be metabolized by other tissues from scratch.
If an equilibrium exists between pyruvate and lactate, and lactate formation is favored in non-Kvothean cells, they need to have means to recover pyruvate. The oxidative component is overlooked, but lactate dehydrogenase occurs in mitochondria for this purpose; the controversy lies in where.
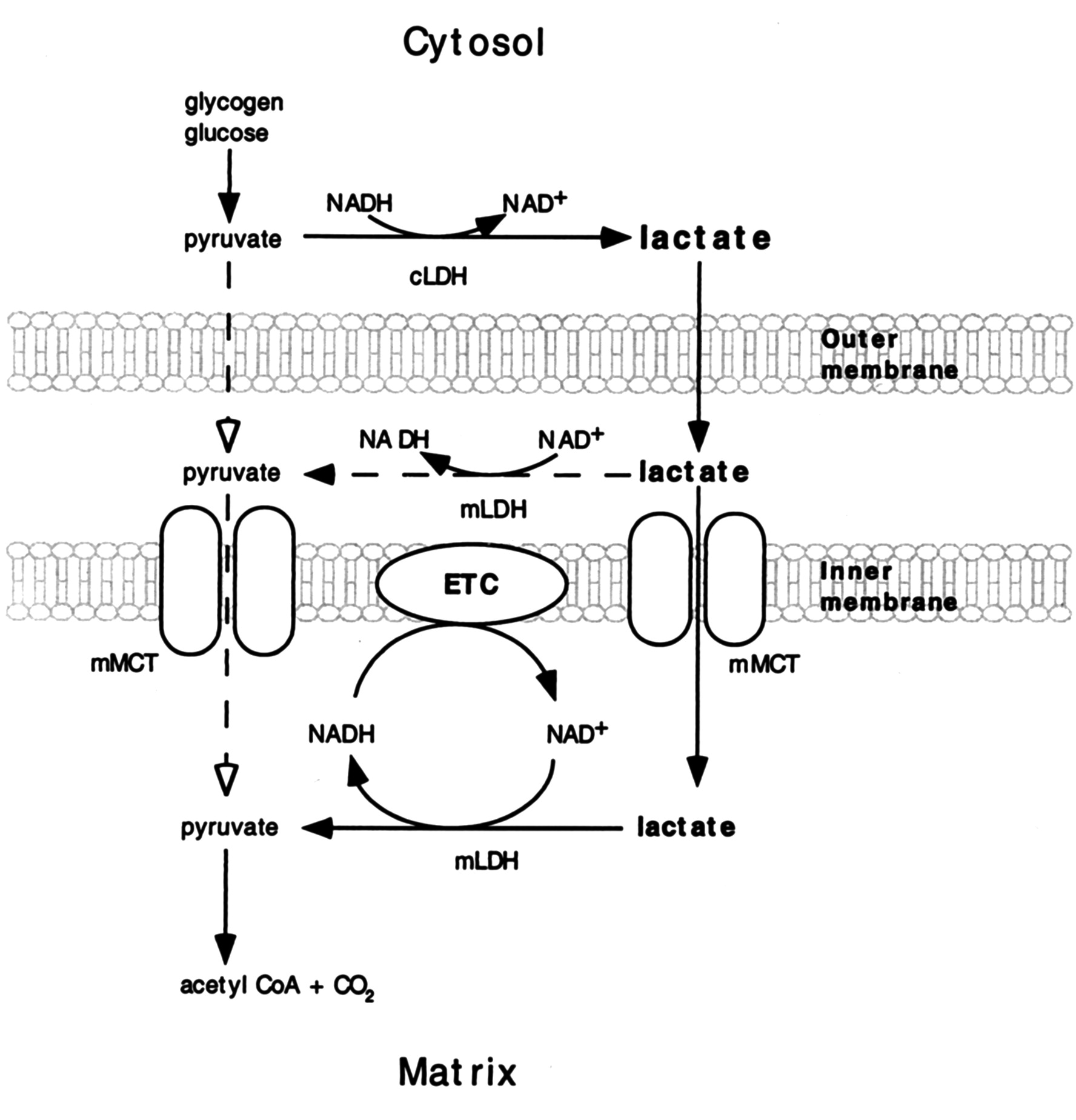
It may be present:
- On the outer surface of the inner membrane (mLDH in the middle, positioned similarly to mGPDH shown earlier); or
- In the matrix (mLDH at the bottom).
The benefit of it occurring in the matrix would be to function as another mitochondrial shuttle, which would take advantage of the pyruvate-lactate distribution with preferential lactate formation. However, the same distribution tendency can be problematic in the matrix for diverting pyruvate and NADH away from their expected fate when levels rise from origins other than lactate oxidation. Lactate dehydrogenase would tend to metabolize them into lactate and NAD+ in place of pyruvate dehydrogenase (PDH) and NADH dehydrogenase (Complex I).
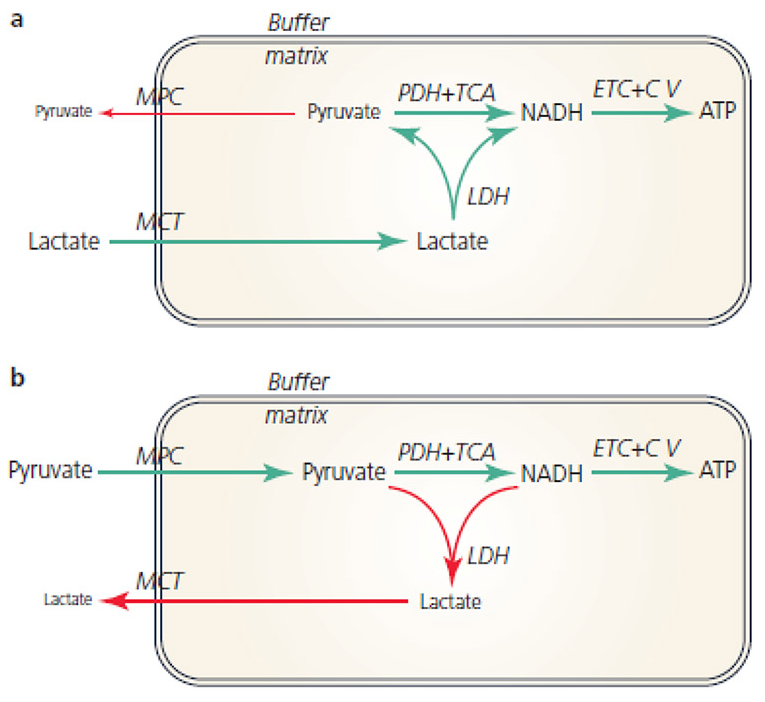
Recall that the equilibrium between lactate and pyruvate is also dictated by NAD+ and NADH. Considering how many sources of NADH there are in the matrix, it would be concerning to have LDH competing for its oxidation.
This is a situation where the different (iso)forms of lactate dehydrogenase would be of help, so that the enzyme had a specific function:
- Lactate → pyruvate: Ach, ja.
- Pyruvate → lactate: Nicht, nicht.
But it doesn't seem to be the case and the location of mitochondrial LDH is deemed to be outside of the matrix to avoid these issues:
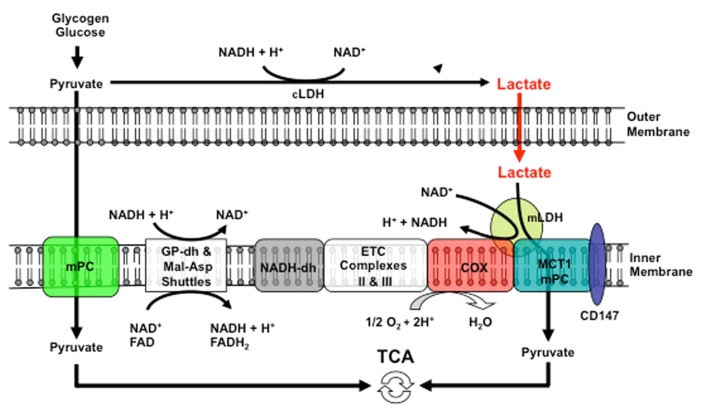
-
Coenzyme Q-cytochrome c oxidoreductase (Complex III) | Wikipedia
↳ Q cycle | WikipediaI thought that improvising a stepwise animation would clarify the cycle better than text. The transfer molecules towards cytochrome c are not shown.

Kekel is a harsh critic of the conventional model, but understanding both sides helps to form an opinion.
-
The following simplified reactions may give an impression that the hydrogens accepted by ubiquinone (UQ) to become ubiquinol (UQH2) are derived from 'NADH + H+' or 'FADH2'.
- [NADH] + H+ → [NAD+ + H–] + H+
- NADH + H+ → NAD+ + (H– + H+)
- NADH + H+ → NAD+ + (2H)
⠀ - FADH2 → FAD + (2H)
⠀ - UQ + 2H → UQH2
The first issue is that the lone proton released by dehydrogenases in the production of NADH is lost to the medium. When NADH reaches complex I to be oxidized, it won't have its original pairing proton, and it can only donate the incorporated hydrogen.
FMN in Complex I accepts it as a hydride ion (H– ⇄ H+ + 2e–) from NADH:
- NADH + FMN → NAD+ + FMNH–
Even though this hydrogen could land on ubi(semi)quinone after a series of reactions, it doesn't seem to.
You may focus on the dark blue to track the proton and red to track electrons, knowing that the top of the rectangle is NAD and the bottom is FMN. Note that when NAD dissociates, the rectangle only shows FMN. The proton is eventually released into the medium.
Protons appear to enter respiratory complexes laterally, near the innermost surface, as electrons are channeled.
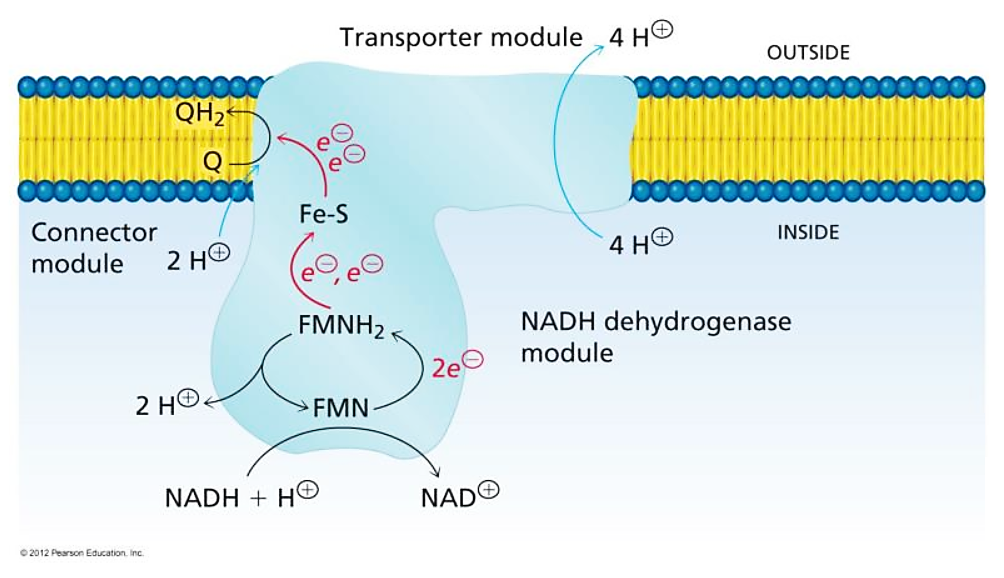
More specifically:
Water route for proton pumping in mitochondrial complex I
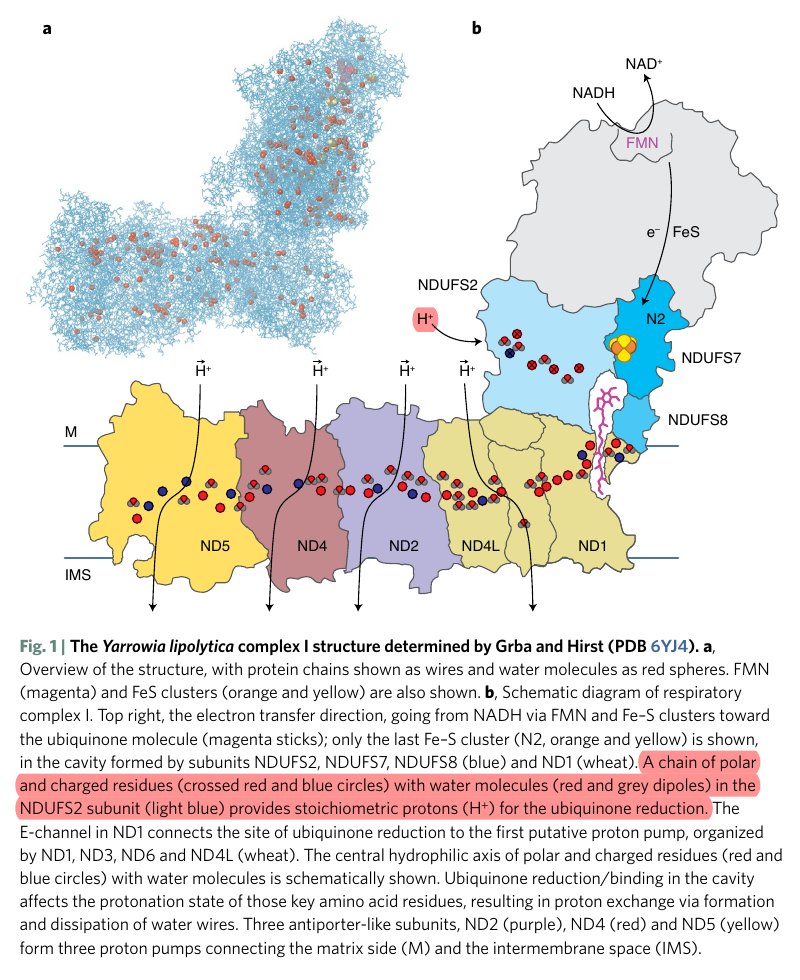
Therefore, the protons liberated in the oxidation of NADH and FADH2 can reach ubiquinone, but likely indirectly when they add to the pool of protons in the matrix. Since the number of free protons is low, they might be buffered or metabolized as fast as they appear.
-
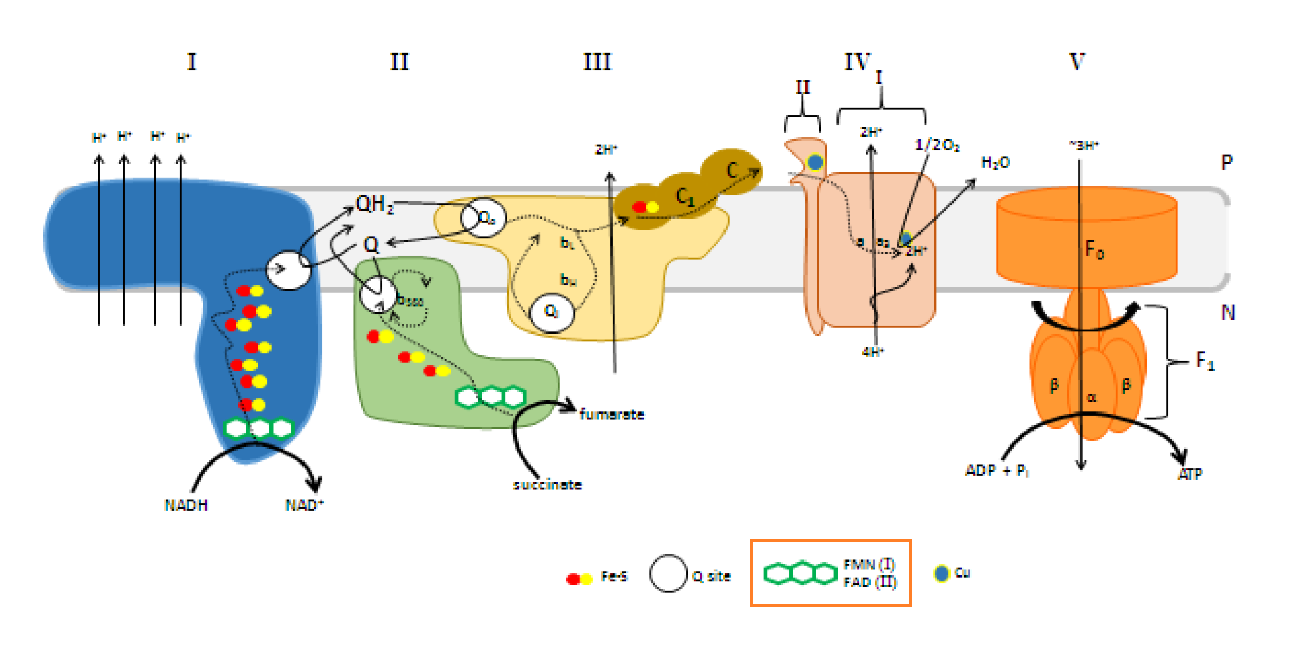
A summary of proton movement in oxidative phosphorylation:
Matrix IMS IMS Matrix → ← 2H⁺  + 4H⁺ →
+ 4H⁺ →Complex I 4H⁺ Complex II  2H⁺
2H⁺2H⁺ 
 2H⁺
2H⁺2H⁺ → Complex III 4H⁺ 4H⁺ Complex III ← 2H⁺ 2H⁺  + 2H⁺ →
+ 2H⁺ →Complex IV 2H⁺ 2H⁺ Complex IV ← 2H⁺ +  2H⁺
2H⁺← → H₂O ← ½O₂ + 2H ↲ ↳ 2H + ½O₂ → H₂O 10H⁺ ← Complex V 10H⁺ 6H⁺ Complex V → 6H⁺ The arrows weren't supposed to be stylized.
-
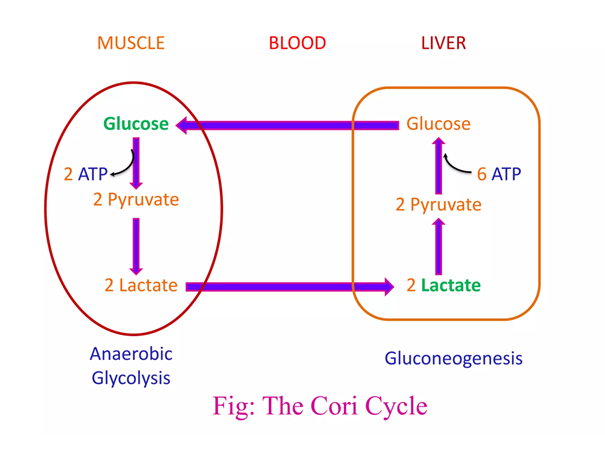
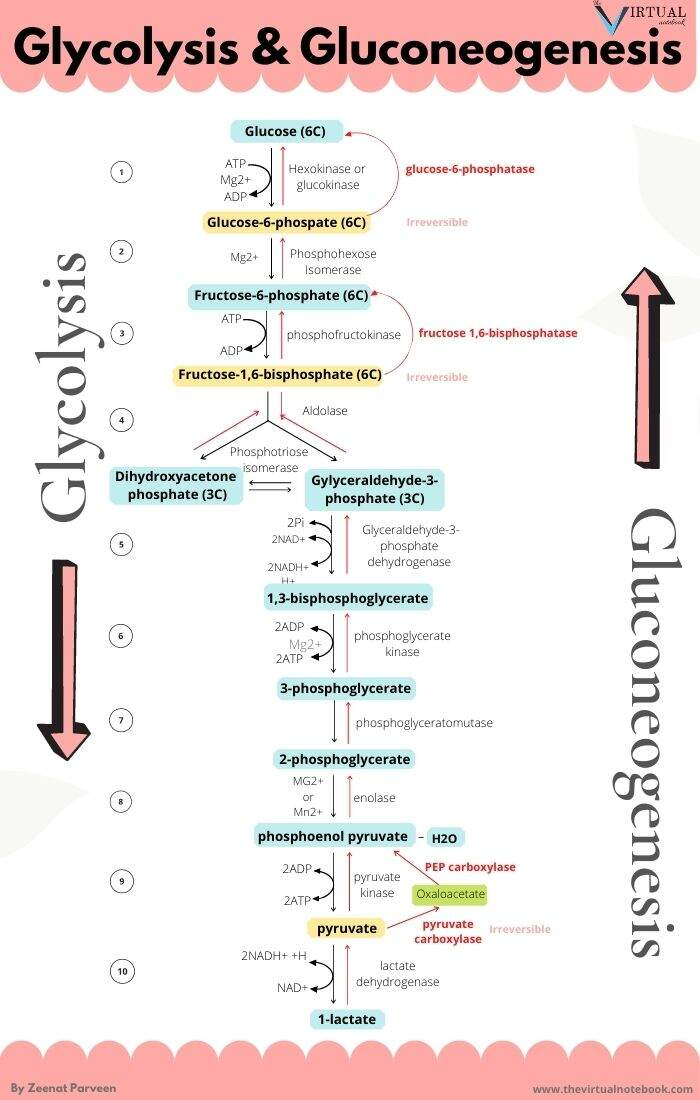
The metabolic steps responsible for the ATP difference in the breakdown and synthesis of dextructose:
Glycolysis (↓) Enzyme Gluconeogenesis (↑) –1 ATP HK N/A–1 ATP PFK N/A+2 ATP PGK –2 ATP +2 ATP PK N/AN/APEPCK –2 ATP (⇄ 2 GTP) N/APC –2 ATP Total Total +2 ATP (–4 ATP/↺) –6 ATP With the exception of PGK, these are the irreversible steps; the others not included are reversible.
If forming and unforming lactate occurred in the same cell, it would have a neutral effect. However, for pyruvate to reappear elsewhere, NADH is consumed in the origin and NAD⁺ in the destination.
-
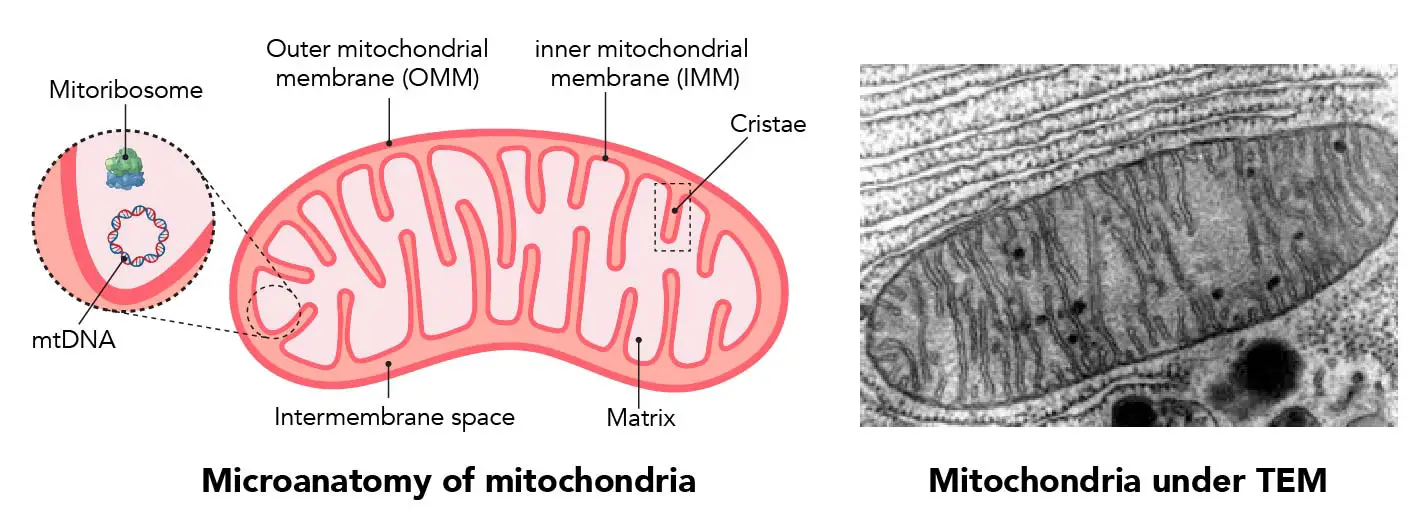
"The IMM consists of subcompartments called cristae and inner boundary membrane (IBM) (Palade, 1953). Cristae are invaginations protruding into the mitochondrial matrix, whereas the IBM runs parallel to the outer mitochondrial membrane (OMM). Cristae and IBM are connected via narrow tubular or slit‐like structures, known as crista junctions (CJs). In recent years, studies show that components of the electron transport chain (ETC) are confined to the lateral surfaces of the cristae rather than equally distributed along the IMM (Vogel et al, 2006; Wilkens et al, 2013). Moreover, dimers of F1F0 ATP Synthase assemble in rows along the edges of the cristae (Dudkina et al, 2005; Strauss et al, 2008; Davies et al, 2011). The CJs can be kept in a closed state by oligomers of the inner‐membrane dynamin‐like GTPase, OPA1 (Frezza et al, 2006; Pham et al, 2016), as well as components of the mitochondrial contact site and cristae organizing system (MICOS complex) (John et al, 2005; Rabl et al, 2009; Barrera et al, 2016; Glytsou et al, 2016)."
"Measurement of ΔΨm in individual cristae reveals that crista junctions provide electrical insulation and sustain polarization of individual mitochondrial cristae within a single mitochondrion even when neighbouring cristae are damaged.
- Cristae have higher ΔΨ compared to their adjoining inner mitochondrial membranes.
- Cristae are electrically insulated, allowing individual cristae within any given mitochondrion to have different membrane potentials.
- Cristae can remain polarized despite depolarization of neighbouring ones.
- Disruption of crista junctions impairs the electrical insulation of cristae, equilibrating their ΔΨ with those of inner mitochondrial membranes."
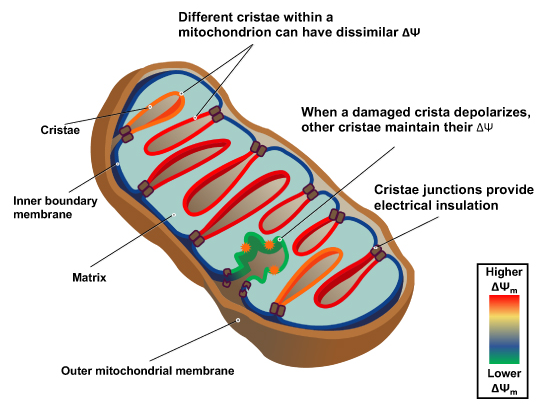
"This study raises interesting questions as to why mitochondria organize the ΔΨm in this way. One advantage, for example, could be related to the fact that ΔΨm constitutes the main energy available to drive protons through F1F0 ATP Synthase to produce ATP. As such, the localization of F1F0 ATP Synthase at the cristae rims appears to be advantageous in terms of proximity to the batteries. Another possible advantage could be compartmentalization of ΔΨm in each crista may serve as a safeguard mechanism restricting the impact of localized damage. In the case of the equipotential model, where the inner membrane of the entire mitochondrion represents a single capacitor, a breach in membrane integrity in one crista would cause a collapse in voltage in all cristae and compromise the function of the whole organelle. If, on the other hand, the IMM could maintain numerous, discrete electrochemical gradients, like a group of batteries, then failure of one or more would not invariably jeopardize the entire mitochondrion. This may be of particular relevance in cells harboring a highly interconnected mitochondrial network as opposed to cells with less elongated and/or branched mitochondria. Furthermore, the hetero‐potential model suggests that cristae with higher ΔΨCr‐IBM could compensate for cristae with impaired function."
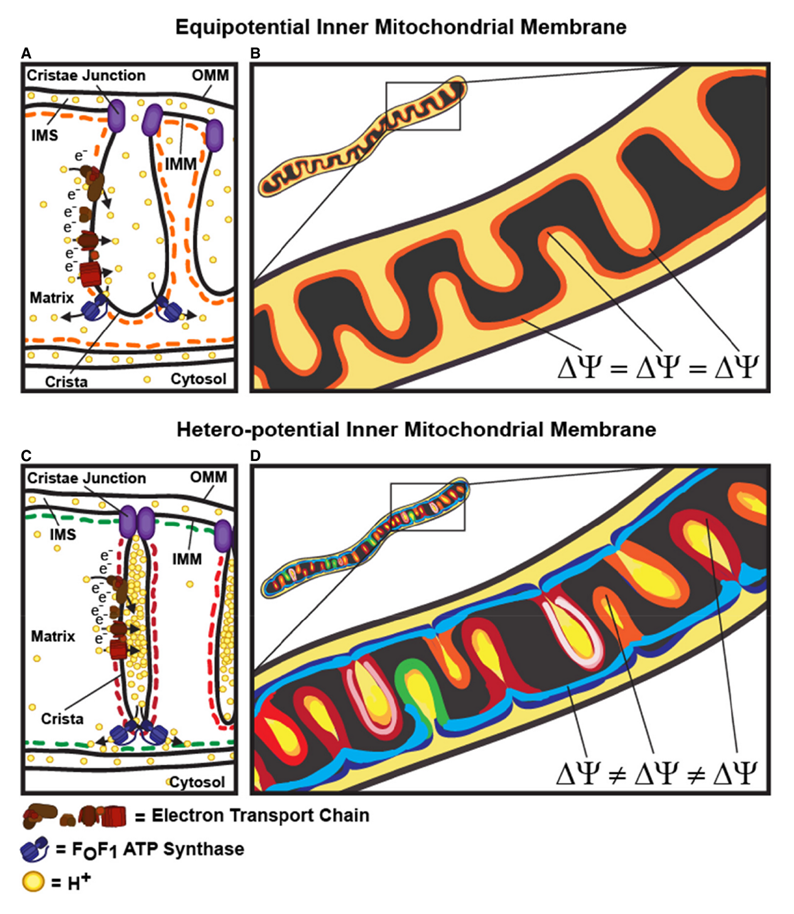
"Hyperpolarized and depolarized IMM potentials are associated with different states of respiration. While both uncoupling and an increased rate of ATP synthesis dissipate ΔΨm, a decrease in ATP synthesis may result in hyperpolarization and increased ROS production. The hetero‐potential model of the mitochondrion allows for different cristae to serve different functions. In this model, some cristae could be more dedicated to ATP synthesis, whereas neighboring cristae could play a role in ROS signaling. The hetero‐potential model further allows for the consideration that different cristae may engage in primarily complex II vs. complex I respiration, which are associated with different membrane potentials and could be driven by different fuels."
Protonic Capacitor: Elucidating the biological significance of mitochondrial cristae formation
"[..]excess protons (positive charges) in an aqueous liquid on one side of a membrane will repel each other to become electrostatically localized along the membrane surface, attracting an equal number of excess hydroxyl anions (negative charges) to the other side of the membrane and thus resulting in a “protonic capacitor structure” (Fig. 2)."
Consequences of Folding the Mitochondrial Inner Membrane
"The greater the energy requirements of a cell, the more inner membrane surface area it contains. Because there are practical limits to the volume fraction that cells can reserve for mitochondria, crista packing is maximized where energy demand is greatest, e.g., in cardiomyocytes the surface area of the inner membrane is more than tenfold that of the outer membrane."
"Although internalizing the chemiosmotic membrane is essential for mass production of ATP, it creates a complex and potentially risky situation for the cell. In particular, conditions that swell the matrix will cause the inner membrane to unfold and, eventually, rupture the outer membrane. In fact, cells use this demolition mechanism when death is the intended outcome. For example, inner membrane “herniation” of the outer membrane is observed in late stages of programmed cell death (extrinsic apoptosis) in FAS-activated liver (
Figure 1). Crista contents, including cytochrome c, spill into the cytosol, resulting in irreversible loss of membrane potential and ATP production (Mootha et al., 2001). Matrix swelling in this case was attributed (Feldmann et al., 2000) to the mitochondrial permeability transition pore, MPTP, the opening of which can drive an osmotic influx of water sufficient to unfold the inner membrane and rupture the outer membrane (e.g., Rasola and Bernardi, 2011).""Extreme crista swelling is as perilous to the cell as uncontrolled matrix swelling, e.g., the total volume of a few fully expanded cristae in a single muscle mitochondrion easily exceeds the volume enclosed by the outer membrane. In fact, rupture of the outer membrane by crista (not matrix) swelling occurs in insect flight muscle as a prelude to apoptosis (Walker and Benzer, 2004). Clearly, the process of unfolding the inner membrane is as important to cell survival as generating the crista folds and likely is regulated as carefully."
"Although, at first glance, it seems risky to fold a large membrane within an outer membrane, rupture of which is fatal, this situation actually provides the cell an advantage. When mitochondria are suspended in hypo-osmotic media, outer membranes lyse at sucrose gradients tenfold greater than liposomes or mitochondrial inner membrane vesicles of similar size, typically 20–30 mM (Douce et al., 1972; Li et al., 1986). This dramatic protection against osmotic stress directly accrues from the outer membrane being osmotically inactive, i.e., very permeable to small solutes. The chemiosmotic inner membrane is the mitochondrial osmometer. Swelling of the matrix caused by osmotic influx of water compresses the cristae before significant pressure is applied to the outer membrane by outward expansion of the inner membrane. In effect, unfolding the inner membrane absorbs significant osmotic stress and delays irreversible damage to the mitochondria. Equally important, this indirect rupture mechanism provides the cell the opportunity to regulate outer membrane lysis."
"A mechanism has been proposed that would protect mitochondria against outer membrane lysis and inner-membrane domain mixing during crista swelling: fusion of tubular cristae to form larger cristae more adaptable to volume changes. Crista fusion was suggested by the first EM tomograms of mammalian mitochondria, which revealed complex cristae with tubular and lamellar regions (Mannella et al., 1997; Perkins et al., 1997). Larger cristae are more prevalent in condensed mitochondria; decreased matrix volume brings cristae into closer proximity, favoring fusion (Mannella et al., 2001). It is likely that crista fusion in response to matrix contraction is quite extensive. Condensed liver mitochondria have large dilated cristae with multiple (up to seven) junctions (Mannella et al., 1997) and condensed yeast mitochondria may have a single dilated internal cavity with much of the inner membrane pulled away from the outer membrane and no well-defined crista junctions or cristae (Mannella et al., 2001)."
-
Hydrogen economy in glycolysis (the charges may be ignored):
Out (–) In (+) HK H⁺ PFK H⁺ 2GAPDH *2H⁻ 2H⁺ 2ENO 2H₂O 2PK 2H⁺ Total 10H 2H (*2H⁻ from 2NADH)
The expectation (–10H + 2H = –8H) is different from the actual change (–6H), when we obtain the pyruvates:
Toxin Formula Glucose C6H12O6 (–8H →) Expected C6H4O6 (–6H →) 2Pyruvate C6H6O6 2Lactate C6H10O6 2Lactic acid C6H12O6 What explains the difference (between expected and actual change) is that the 2H⁺ released at the dehydrogenase level (GAPDH) are derived from inorganic phosphate rather than extracted from the glucose metabolite (
NADH + H⁺). When we disconsider these 2H⁺, we arrive on the –6H needed.
As for lactate, when it's formed, 4H are consumed from 2H⁻ + 2H⁺. Going backwards, it would be equivalent to never having produced:
- H⁺ (HK)
- H⁺ (PFK)
- 2H⁻ (2GAPDH)
Then, we're left with –2H for 2 lactates (C6H10O6) to match with a glucose molecule (C6H12O6).
Even though 2H⁺ were disconsidered previously, they still occur, just not extracted from glucose metabolites. After canceling out the 3 products listed above, we get:
Out (–) In (+) 2GAPDH 2H⁺ 2ENO 2H₂O 2PK 2H⁺ Total 6H 2H When lactate is exported from the cell, it tends to bring a H⁺ along as counter-ion. So, 2 lactates would take out the equivalent to those 2H⁺ produced at 2GAPDH.
What remains is the bottom of the original table:
- 2H₂O out (produced)
- 2H⁺ in (consumed)
Therefore, lactate synthesis and export is an alkalinizing process for the cell:
- H⁺ are consumed when pyruvates are formed (pyruvate enol-phosphate + H⁺ → pyruvate), to then become lactates to be exported. In contrast, the H₂O molecules produced are neutral.
- LDH reaction consumes H⁺ directly (pyruvate + NADH + H⁺ → lactate + NAD⁺)
- Additional H⁺ tend to be eliminated with lactates as counter-ions (lactate + H⁺)
Nevertheless, lactate is associated with acidification for a few reasons that I'm aware of:
The coexport with a H⁺ alkalinizes the interior and acidifies the exterior of the cell. It's easier to monitor the outside, so that's what we associate with.
Once this lactic acid equivalent leaves the cell, the H⁺ is quickly buffered, leaving lactate to pair with available cations, such as Na⁺ ('HLac' + NaHCO3 → NaLac + H2CO3). If not by using up hydrocarbonate ions, a surge of lactate with expansion of the pool of organic anions can lead to an increase in cations to maintain ion neutrality. This tends to reflect on the concentration of free H⁺, that may rise in proportion along.
Marked ATP hydrolysis, that often coincides with excess lactate formation, is acidifying.
- ATP + H₂O → ADP + Pi + H⁺
Rather than the theoretical source from the previous section, this H⁺ can be the pair for lactate.
The following diagram has these events simplified:
Metabolic acidosis and fatigue: Where to from here?
-
Observe the inconsistency in the conventional nomenclature for glycolysis metabolites:
Abbreviation Metabolite GLC Glucose G6P Glucose-6-phosphate F6P Fructose-6-phosphate F1,6BP Fructose-1,6-bisphosphate DHAP Dihydroxyacetone-3-phosphate G3P Glyceraldehyde-3-phosphate 1,3BPG 1,3-Bisphosphoglycerate 3PG 3-Phosphoglycerate 2PG 2-Phosphoglycerate PEP Phosphoenolpyruvate PYR Pyruvate I think that they put phosphate in evidence when the molecule becomes a phosphate donor, similar to creatine (phosphocreatine, which is creatine phosphate), but to relegate the core is confusing, and it's worse without bolding.
Standardizing against the norm makes it easier to grasp:
Abbreviation Metabolite GLC Glucose G6P Glucose 6-phosphate F6P Fructose 6-phosphate F1,6BP Fructose 1,6-bisphosphate DHAP Dihydroxyacetone 3-phosphate G3P Glyceral 3-phosphate 1,3BPG Glyceral 1,3-bisphosphate 3PG Glycerate 3-phosphate 2PG Glycerate 2-phosphate PEP Pyruvate Enol-phosphate PYR Pyruvate The numbers next to phosphate indicate the carbon position where phosphates are attached to the molecule.
- For a molecule with 6 carbons, we can infer that a '6-phosphate' is a a phosphate attached to one extremity.
- When we get '1,6-bisphosphate', the molecule contains phosphates on both extremities.
In working with split molecules of 3 carbons, the same approach applies.
- Bis- and tris-: separate (example: P-Molecule-P)
- Di- and tri-: in sequence (example: Molecule-PP)
High-energy phosphate | Wikipedia
"Often, high-energy phosphate bonds are denoted by the character '~'. In this "squiggle" notation, ATP becomes A-P~P~P. The squiggle notation was invented by Fritz Albert Lipmann, who first proposed ATP as the main energy transfer molecule of the cell, in 1941. Lipmann's notation emphasizes the special nature of these bonds. Stryer states:
ATP is often called a high energy compound and its phosphoanhydride bonds are referred to as high-energy bonds. There is nothing special about the bonds themselves. They are high-energy bonds in the sense that free energy is released when they are hydrolyzed, for the reasons given above. Lipmann's term "high-energy bond" and his symbol ~P (squiggle P) for a compound having a high phosphate group transfer potential are vivid, concise, and useful notations. In fact Lipmann's squiggle did much to stimulate interest in bioenergetics."
Number of carbons Saccharide (-ose*) 6 Hexose 5 Pentose 4 Tetrose 3 Triose 2Diose1Monose*As in Alberto's 'godnose'.
The upper part of the glycolysis list are the hexoses (until F1,6BP). After splitting (below F1,6BP), we get the trioses. A hexose or triose phosphate is self-explanatory.
When glucose (6-phosphate) is diverted from glycolysis for nucleotide synthesis, it undergoes oxidative decrapoxylation (losing a carbon), becoming a pentose. It gives name to the Pentose Phosphate Pathway (sometimes referred to as Hexose Monophosphate Shunt).